Hydraulic Fracturing in the Toughness-Dominated Regime
Hydraulic Fracturing in the Toughness-Dominated Regime
Under the toughness-dominated regime without leak-off, the fluid viscosity dissipation is negligible compared to the energy released for fracture surface formation (Detournay et al., 2016). Therefore, in this regime, we can neglect the pressure loss within the fracture and use the Sneddon solution for crack opening (Bourdin et al., 2012, Sneddon et al., 1969) to determine the pressure and length evolution.
The work of the pressure force is as follows:
$$ \begin{equation} \mathcal{W}(R) =\frac{2 p^2 a^2}{E'}. \end{equation} $$Applying Clapeyron’s theorem, the elastic energy can be determined as
$$ \begin{equation} \mathcal{E}(R) =-\frac{\pi p^2 a^2}{E'}, \end{equation} $$and the energy release rate with respect to the crack length $a$ propagating along the initial inclination is
$$ \begin{equation} G(R) = - \frac{\partial \mathcal{E}}{\partial (2 a)} = \frac{\pi p^2 a}{E'}. \end{equation} $$Griffith's criterion (Griffith et al., 1920) states that the fracture propagates when $G=G_c$ and the critical volume for crack propagation is $V_c := \sqrt{\dfrac{4 \pi G_c a^3}{E' }}$ in a quasi-static volume control setting (the fracture propagation is always unstable with pressure control).
The evolution of the corresponding pressure and fracture length
$$ \begin{equation} p(V) = \begin{cases} \dfrac{E' V}{2 \pi a_0^2} &\text{for} V < V_c \\ \left[ \dfrac{2 E' G_c^2}{\pi V} \right] ^{\frac{1}{3}}&\text{for} V \geq V_c, \end{cases} \end{equation} $$ $$ \begin{equation} a(V) = \begin{cases} a_0 & V < V_c \\ \left[ \dfrac{E' V^2}{4 \pi G_c} \right ] ^{\frac{1}{3}} & V \geq V_c. \end{cases} \end{equation} $$Problem Description
Based on Sneddon's solution (Sneddon et al., 1969), we verified the model with plane-strain hydraulic fracture propagation in a toughness-dominated regime. In an infinite 2D domain, the problem was solved with a line fracture $[-a_0, a_0] \times \{0\}$ ($a_0$ = 0.05). We considered a large finite domain $[-40a_0,40a_0] \times [-40a_0,40a_0]$ to account for the infinite boundaries in the closed-form solution. The effective size of an element, $h$, is $1\times10^{-2}$.

- In order to have the hydraulic fracturing in the toughness dominated-regime, add, $\texttt{
propagating }$ in the project file. - Yoshioka et al., 2019 provides additional information on the implementation, use of real material properties, and rescaling of the phase-field energy functional.
Input Data
Taking advantage of the linearity of the system, the simulations were run with the dimensionless properties listed in the Table below.
| Name | Value | Symbol |
|---|---|---|
| Young’s modulus | 1 | $E$ |
| Poisson’s ratio | 0.15 | $\nu$ |
| Fracture toughness | 1 | $G_{c}$ |
| Regularization parameter | 2$h$ | $\ell_s$ |
| Length | 4 | $L$ |
| Height | 4 | $H$ |
| Initial crack length | 0.1 | $2a_0$ |
import argparse
import math
import os
import re
import time
import gmsh
import matplotlib.pyplot as plt
import numpy as np
from ogs6py import ogs
from ogstools.msh2vtu import run
pi = math.pi
plt.rcParams["text.usetex"] = TrueE = 1.0
nu = 0.15
Gc = 1.0
h = 0.01
a0 = 0.05 # half of the initial crack length
phasefield_model = "AT1" # AT1/AT2Output Directory and Project File
# file's name
prj_name = "Kregime_Propagating.prj"
meshname = "mesh_full_pf"
from pathlib import Path
out_dir = Path(os.environ.get("OGS_TESTRUNNER_OUT_DIR", "_out"))
if not out_dir.exists():
out_dir.mkdir(parents=True)Mesh Generation
def mesh_generation(lc, lc_fine):
"""
lc ... characteristic length for coarse meshing
lc_fine ... characteristic length for fine meshing
"""
L = 4.0 # Length
H = 4.0 # Height
b = 0.4 # Length/Height of subdomain with fine mesh
# Before using any functions in the Python API, Gmsh must be initialized
gmsh.initialize()
gmsh.option.setNumber("General.Terminal", 1)
gmsh.model.add("rectangle")
# Dimensions
dim0 = 0
dim1 = 1
dim2 = 2
# Points
gmsh.model.geo.addPoint(-L / 2, -H / 2, 0, lc, 1)
gmsh.model.geo.addPoint(L / 2, -H / 2, 0, lc, 2)
gmsh.model.geo.addPoint(L / 2, H / 2, 0, lc, 3)
gmsh.model.geo.addPoint(-L / 2, H / 2, 0, lc, 4)
gmsh.model.geo.addPoint(-b, -b - lc_fine / 2, 0, lc_fine, 5)
gmsh.model.geo.addPoint(b, -b - lc_fine / 2, 0, lc_fine, 6)
gmsh.model.geo.addPoint(b, b + lc_fine / 2, 0, lc_fine, 7)
gmsh.model.geo.addPoint(-b, b + lc_fine / 2, 0, lc_fine, 8)
# Lines
gmsh.model.geo.addLine(1, 2, 1)
gmsh.model.geo.addLine(2, 3, 2)
gmsh.model.geo.addLine(3, 4, 3)
gmsh.model.geo.addLine(4, 1, 4)
gmsh.model.geo.addLine(5, 6, 5)
gmsh.model.geo.addLine(6, 7, 6)
gmsh.model.geo.addLine(7, 8, 7)
gmsh.model.geo.addLine(8, 5, 8)
# Line loops
gmsh.model.geo.addCurveLoop([1, 2, 3, 4], 1)
gmsh.model.geo.addCurveLoop([5, 6, 7, 8], 2)
# Add plane surfaces defined by one or more curve loops.
gmsh.model.geo.addPlaneSurface([1, 2], 1)
gmsh.model.geo.addPlaneSurface([2], 2)
gmsh.model.geo.synchronize()
# Prepare structured grid
gmsh.model.geo.mesh.setTransfiniteCurve(
6, math.ceil(2 * b / lc_fine + 2), "Progression", 1
)
gmsh.model.geo.mesh.setTransfiniteCurve(
8, math.ceil(2 * b / lc_fine + 2), "Progression", 1
)
gmsh.model.geo.mesh.setTransfiniteSurface(2, "Alternate")
gmsh.model.geo.mesh.setRecombine(dim2, 1)
gmsh.model.geo.mesh.setRecombine(dim2, 2)
gmsh.model.geo.synchronize()
# Physical groups
P1 = gmsh.model.addPhysicalGroup(dim0, [1])
gmsh.model.setPhysicalName(dim0, P1, "P1")
P2 = gmsh.model.addPhysicalGroup(dim0, [2])
gmsh.model.setPhysicalName(dim0, P2, "P2")
Bottom = gmsh.model.addPhysicalGroup(dim1, [1])
gmsh.model.setPhysicalName(dim1, Bottom, "Bottom")
Right = gmsh.model.addPhysicalGroup(dim1, [2])
gmsh.model.setPhysicalName(dim1, Right, "Right")
Top = gmsh.model.addPhysicalGroup(dim1, [3])
gmsh.model.setPhysicalName(dim1, Top, "Top")
Left = gmsh.model.addPhysicalGroup(dim1, [4])
gmsh.model.setPhysicalName(dim1, Left, "Left")
Computational_domain = gmsh.model.addPhysicalGroup(dim2, [1, 2])
gmsh.model.setPhysicalName(dim2, Computational_domain, "Computational_domain")
gmsh.model.geo.synchronize()
output_file = f"{out_dir}/" + meshname + ".msh"
gmsh.model.mesh.generate(dim2)
gmsh.write(output_file)
gmsh.finalize()Pre-Processing
def pre_processing(h, a0):
mesh = pv.read(f"{out_dir}/mesh_full_pf_domain.vtu")
phase_field = np.ones((len(mesh.points), 1))
pv.set_plot_theme("document")
for node_id, x in enumerate(mesh.points):
if (
(mesh.center[0] - x[0]) <= a0 + 0.001 * h
and (mesh.center[0] - x[0]) >= -a0 - 0.001 * h
and (mesh.center[1] - x[1]) < h / 2 + 0.001 * h
and (mesh.center[1] - x[1]) > -h / 2 - 0.001 * h
):
phase_field[node_id] = 0.0
mesh.point_data["phase-field"] = phase_field
mesh.save(f"{out_dir}/mesh_full_pf_OGS_pf_ic.vtu")Run the Simulation
import pyvista as pv
pv.set_plot_theme("document")
if "PYVISTA_HEADLESS" in os.environ:
pv.start_xvfb()
pv.set_jupyter_backend("static")
def Hydraulic_Fracturing_Toughness_Dominated_numerical(h, phasefield_model):
# mesh properties
ls = 2 * h
# generate prefix from properties
filename = f"results_h_{h:0.4f}_{phasefield_model}"
mesh_generation(0.1, h)
# Convert GMSH (.msh) meshes to VTU meshes appropriate for OGS simulation.
input_file = f"{out_dir}/" + meshname + ".msh"
args = argparse.Namespace(
filename=input_file,
output=f"{out_dir}/" + meshname,
dim=0,
delz=False,
swapxy=False,
rdcd=True,
ogs=True,
ascii=False,
)
run(args)
# As a preprocessing step, define the initial phase-field (crack).
pre_processing(h, a0)
# change properties in prj file #For more information visit: https://github.com/joergbuchwald/ogs6py
model = ogs.OGS(
INPUT_FILE=prj_name,
PROJECT_FILE=f"{out_dir}/{prj_name}",
MKL=True,
args=f"-o {out_dir}",
)
model.replace_parameter_value(name="ls", value=ls)
model.replace_text(phasefield_model, xpath="./processes/process/phasefield_model")
model.replace_text(filename, xpath="./time_loop/output/prefix")
model.write_input()
# run simulation with ogs
t0 = time.time()
print(">>> OGS started execution ... <<<")
! ogs {out_dir}/{prj_name} -o {out_dir} > {out_dir}/log.txt
tf = time.time()
print(">>> OGS terminated execution <<< Elapsed time: ", round(tf - t0, 2), " s.")Hydraulic_Fracturing_Toughness_Dominated_numerical(h, phasefield_model)Info : Meshing 1D...
Info : [ 0%] Meshing curve 1 (Line)
Info : [ 20%] Meshing curve 2 (Line)
Info : [ 30%] Meshing curve 3 (Line)
Info : [ 40%] Meshing curve 4 (Line)
Info : [ 60%] Meshing curve 5 (Line)
Info : [ 70%] Meshing curve 6 (Line)
Info : [ 80%] Meshing curve 7 (Line)
Info : [ 90%] Meshing curve 8 (Line)
Info : Done meshing 1D (Wall 0.000515956s, CPU 0.000756s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 1 (Plane, Frontal-Delaunay)
Info : [ 0%] Blossom: 24032 internal 482 closed
Info : [ 0%] Blossom recombination completed (Wall 0.314673s, CPU 0.309132s): 7878 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.793024, min Q = 0.371315
Info : [ 60%] Meshing surface 2 (Transfinite)
Info : Done meshing 2D (Wall 0.467246s, CPU 0.461732s)
Info : 14439 nodes 14848 elements
Info : Writing '/var/lib/gitlab-runner/builds/vZ6vnZiU/0/ogs/build/release-petsc/Tests/Data/PhaseField/Kregime_Propagating_jupyter_notebook/Kregime_Propagating_jupyter/mesh_full_pf.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/vZ6vnZiU/0/ogs/build/release-petsc/Tests/Data/PhaseField/Kregime_Propagating_jupyter_notebook/Kregime_Propagating_jupyter/mesh_full_pf.msh'
Output: /var/lib/gitlab-runner/builds/vZ6vnZiU/0/ogs/build/release-petsc/Tests/Data/PhaseField/Kregime_Propagating_jupyter_notebook/Kregime_Propagating_jupyter/mesh_full_pf
Original mesh (read)
14439 points in 3 dimensions; cells: 2 vertex, 160 line, 14358 quad; point_data=['gmsh:dim_tags']; cell_data=['gmsh:physical', 'gmsh:geometrical']; cell_sets=['P1', 'P2', 'Bottom', 'Right', 'Top', 'Left', 'Computational_domain', 'gmsh:bounding_entities']
##
Detected mesh dimension: 2
##
Domain mesh (written)
14439 points in 3 dimensions; cells: 14358 quad; point_data=['original_node_number']; cell_data=['MaterialIDs']; cell_sets=[]
##
Boundary mesh (written)
160 points in 3 dimensions; cells: 160 line; point_data=['bulk_node_ids']; cell_data=['bulk_elem_ids']; cell_sets=[]
##
Submesh P1 (written)
1 points in 3 dimensions; cells: 1 vertex; point_data=['bulk_node_ids']; cell_data=['gmsh:physical']; cell_sets=[]
##
Submesh P2 (written)
1 points in 3 dimensions; cells: 1 vertex; point_data=['bulk_node_ids']; cell_data=['gmsh:physical']; cell_sets=[]
##
Submesh Bottom (written)
41 points in 3 dimensions; cells: 40 line; point_data=['bulk_node_ids']; cell_data=['bulk_elem_ids']; cell_sets=[]
##
Submesh Right (written)
41 points in 3 dimensions; cells: 40 line; point_data=['bulk_node_ids']; cell_data=['bulk_elem_ids']; cell_sets=[]
##
Submesh Top (written)
41 points in 3 dimensions; cells: 40 line; point_data=['bulk_node_ids']; cell_data=['bulk_elem_ids']; cell_sets=[]
##
Submesh Left (written)
41 points in 3 dimensions; cells: 40 line; point_data=['bulk_node_ids']; cell_data=['bulk_elem_ids']; cell_sets=[]
##
Submesh Computational_domain (written)
14439 points in 3 dimensions; cells: 14358 quad; point_data=['bulk_node_ids']; cell_data=['MaterialIDs']; cell_sets=[]
##
>>> OGS started execution ... <<<
>>> OGS terminated execution <<< Elapsed time: 263.86 s.
Post-Processing
Analytical Solution for the Evolution of Fracture Length and Pressure
def Analytical_solution(phasefield_model, h):
v = np.linspace(1e-10, 0.3, 31)
pres = np.linspace(0, 1.0, 31)
length = np.linspace(0, 1.0, 31)
ls = 2 * h
# Effective Gc and a for AT1/A2
if phasefield_model == "AT1":
Gc_ref = Gc * (3 * h / 8.0 / ls + 1.0)
a_eff = a0 * (1 + pi * ls / (4.0 * a0 * (3 * h / 8.0 / ls + 1.0)))
elif phasefield_model == "AT2":
Gc_ref = Gc * (h / (2.0 * ls) + 1.0)
a_eff = a0 * (1 + pi * ls / (4.0 * a0 * (h / (2.0 * ls) + 1.0)))
Eprime = E / (1 - nu**2)
V_c = (4 * pi * Gc_ref * a_eff**3 / Eprime) ** 0.5
P_c = (Eprime * Gc_ref / (pi * a_eff)) ** 0.5
for i in range(len(v)):
if v[i] < V_c:
pres[i] = Eprime * v[i] / (2 * pi * a_eff**2) / P_c
length[i] = a_eff
else:
pres[i] = (2 * Eprime * Gc_ref**2 / (pi * v[i])) ** 0.333333 / P_c
length[i] = (Eprime * v[i] ** 2 / (4 * pi * Gc_ref)) ** 0.333333
return v, pres, length, Gc_ref, P_c
fluidVolume_analytical = Analytical_solution(phasefield_model, h)[0]
pressure_analytical = Analytical_solution(phasefield_model, h)[1]
length_analytical = Analytical_solution(phasefield_model, h)[2]
Gc_ref = Analytical_solution(phasefield_model, h)[3]
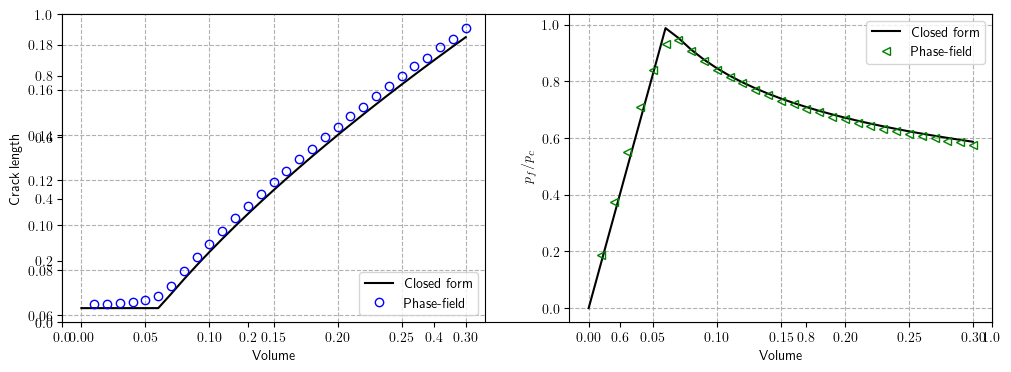
P_c = Analytical_solution(phasefield_model, h)[4]Phase Field Versus Analytical Solution for Fracture Length and Pressure Evolution
In phase field approach, we can retrieve the crack length $a$ as:
$$ \begin{equation} a = \dfrac{\displaystyle \int_\Omega \frac{G_c}{4 c_n} \left(\frac{(1-v)^n}{\ell}+ \ell |\nabla v|^2\right)\,\mathrm{d} \Omega }{G_c \left( \dfrac{h}{4 c_n \ell} + 1 \right)}. \end{equation} $$where $n=1$ corresponds to $\texttt{AT}_1$ ($c_n = 2/3$) and $n=2$ to $\texttt{AT}_2$ ($c_n = 1/2$).
plt.subplots(figsize=(12, 4))
plt.subplot(1, 2, 1)
fluid_volume = []
surface_energy = []
# Open the file for reading
with (out_dir / "log.txt").open() as fd:
# Iterate over the lines
for _i, line in enumerate(fd):
match_surface_energy = re.search(
r"""Surface energy: (\d+\.\d*) Pressure work: (\d+\.\d*) at time: (\d+\.\d*)""",
line,
)
if match_surface_energy:
surface_energy.append(float(match_surface_energy.group(1)))
fluid_volume.append(float(match_surface_energy.group(3)))
plt.grid(linestyle="dashed")
plt.xlabel("Volume")
plt.ylabel("Crack length")
plt.plot(fluidVolume_analytical, length_analytical, "black", label="Closed form")
plt.plot(
fluid_volume,
np.array(surface_energy[:]) / Gc_ref / 2,
"bo",
fillstyle="none",
label="Phase-field",
)
legend = plt.legend(loc="lower right")
plt.subplot(1, 2, 2)
fluid_volume = []
pressure = []
# Open the file for reading
with (out_dir / "log.txt").open() as fd:
# Iterate over the lines
for _i, line in enumerate(fd):
match_pressure = re.search(
r"""Pressure: (\d+\.\d*) at time: (\d+\.\d*)""", line
)
if match_pressure:
fluid_volume.append(float(match_pressure.group(2)))
pressure.append(float(match_pressure.group(1)))
plt.xlabel("Volume")
plt.ylabel("$p_f/p_c$")
plt.plot(fluidVolume_analytical, pressure_analytical, "black", label="Closed form")
plt.plot(
np.array(fluid_volume[:]),
np.array(pressure[:]) / P_c,
"g<",
fillstyle="none",
label="Phase-field",
)
plt.grid(linestyle="dashed")
legend = plt.legend(loc="upper right")
plt.show()
plt.subplots(figsize=(12, 4))
plt.subplot(1, 2, 1)
plt.grid(linestyle="dashed")
plt.plot(
fluidVolume_analytical[1:],
(
abs(length_analytical[1:] - np.array(surface_energy[:]) / (2 * Gc_ref))
/ (np.array(surface_energy[:]) / (2 * Gc_ref))
)
* 100,
"-.ob",
fillstyle="none",
label="Closed form",
)
plt.ylim([0.0, 10])
plt.xlabel("Volume")
plt.ylabel(
r"$\frac{|a_\mathrm{num}-{a}_\mathrm{ana}|}{{a}_\mathrm{num}}\times 100\%$",
fontsize=14,
)
plt.subplot(1, 2, 2)
plt.grid(linestyle="dashed")
plt.plot(
fluidVolume_analytical[1:],
(abs(pressure_analytical[1:] - np.array(pressure[:]) / P_c) / pressure[:]) * 100,
"-.<g",
fillstyle="none",
label="Closed form",
)
plt.ylim([0.0, 10])
plt.xlabel("Volume")
plt.ylabel(
r"$\frac{|p_\mathrm{num}-{p}_\mathrm{ana}|}{{p}_\mathrm{num}}\times 100\%$",
fontsize=14,
)
plt.show()
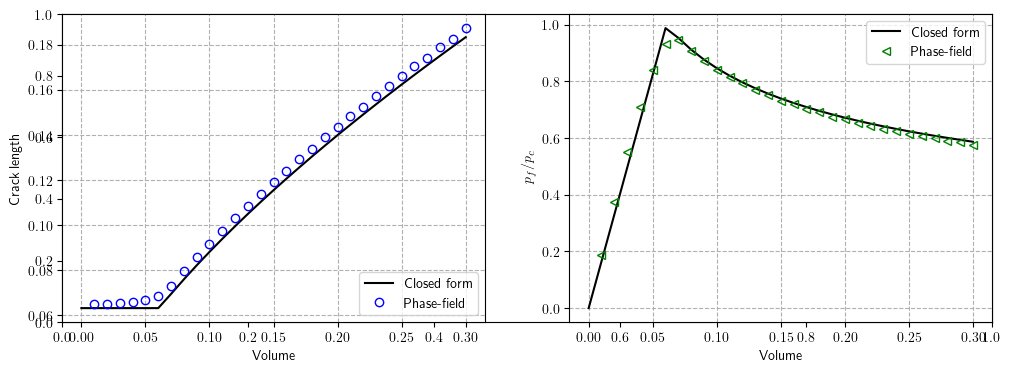
In order to reduce computation time, we perform the simulation with a coarse mesh; the results for the $\texttt{AT}_1$ Model with a mesh size of $h=0.001$ are provided below.
$\texttt{AT}_1$ Model:


Hydraulic Fracturing Animation (Using Phase Field Approach)
import pyvista as pv
pv.set_plot_theme("document")
pv.set_jupyter_backend("static")filename = f"results_h_{h:0.4f}_{phasefield_model}"
reader = pv.get_reader(f"{out_dir}/" + filename + ".pvd")
plotter = pv.Plotter(shape=(1, 2), border=False)
plotter.open_gif(f"{out_dir}/Kregime_Propagating.gif")
for time_value in reader.time_values:
reader.set_active_time_value(time_value)
mesh = reader.read()[0]
sargs = {
"title": "Phase field",
"title_font_size": 16,
"label_font_size": 12,
"n_labels": 5,
"position_x": 0.25,
"position_y": 0.15,
"fmt": "%.1f",
"width": 0.5,
}
p = pv.Plotter(shape=(1, 2), border=False)
clim = [0, 1.0]
points = mesh.point_data["phasefield"].shape[0]
xs = mesh.points[:, 0]
ys = mesh.points[:, 1]
pf = mesh.point_data["phasefield"]
plotter.clear()
plotter.add_mesh(
mesh,
scalars=pf,
show_scalar_bar=True,
colormap="coolwarm",
clim=clim,
scalar_bar_args=sargs,
lighting=False,
)
plotter.add_text(f"Time: {time_value:0.02f}", color="black")
plotter.view_xy()
plotter.write_frame()
plotter.close()DRI3 not available
failed to load driver: zink
Phase Field Profile at Last Time Step
mesh = reader.read()[0]
points = mesh.point_data["phasefield"].shape[0]
xs = mesh.points[:, 0]
ys = mesh.points[:, 1]
pf = mesh.point_data["phasefield"]
clim = [0, 1.0]
sargs = {
"title": "Phase field",
"title_font_size": 16,
"label_font_size": 12,
"n_labels": 5,
"position_x": 0.25,
"position_y": 0.0,
"fmt": "%.1f",
"width": 0.5,
}
plotter = pv.Plotter(shape=(1, 2), border=False)
plotter.add_mesh(
mesh,
scalars=pf,
show_edges=False,
show_scalar_bar=True,
colormap="coolwarm",
clim=clim,
scalar_bar_args=sargs,
lighting=False,
)
plotter.view_xy()
plotter.camera.zoom(1.5)
plotter.window_size = [1000, 500]
plotter.show()DRI3 not available
failed to load driver: zink
References
[1] Detournay, Emmanuel. Mechanics of hydraulic fractures. Annual review of fluid mechanics 48 (2016): 311-339.
[2] Bourdin, Blaise, Chukwudi Chukwudozie, and Keita Yoshioka. A variational approach to the numerical simulation of hydraulic fracturing. In SPE annual technical conference and exhibition. OnePetro, 2012.
[3] Sneddon, Ian Naismith, and Morton Lowengrub. Crack problems in the classical theory of elasticity. 1969, 221 P (1969).
[4] Griffith, Alan Arnold. VI. The phenomena of rupture and flow in solids. Philosophical transactions of the royal society of london. Series A, containing papers of a mathematical or physical character 221, no. 582-593 (1921): 163-198.
[5] Yoshioka, Keita, Francesco Parisio, Dmitri Naumov, Renchao Lu, Olaf Kolditz, and Thomas Nagel. Comparative verification of discrete and smeared numerical approaches for the simulation of hydraulic fracturing. GEM-International Journal on Geomathematics 10, no. 1 (2019): 1-35.
This article was written by Mostafa Mollaali, Keita Yoshioka. If you are missing something or you find an error please let us know.
Generated with Hugo 0.122.0
in CI job 449919
|
Last revision: March 3, 2023

