A 2D GREAT cell benchmark suite simulated using hydro-mechanical variational phase-field process
import os…
(click to toggle)
import os
import sys
from pathlib import Path
from subprocess import run
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import ogstools as ot
import pyvista as pv
from matplotlib import colormaps
mechanics_path = Path(
"..", "..", "LIE", "Mechanics", "GreatCelljupyterNotebook"
).resolve()
sys.path.insert(0, str(mechanics_path))
# Local modules
from mesh_generator import ( # noqa: E402
mesh_GreatCell_intact,
mesh_GreatCell_VPF,
)
from ogs_model_runner import SingleOGSModel # noqa: E402
from Plotter import Plotter # noqa: E402
def truncated_cmap(name, minval=0.3, maxval=0.9, n=6):…
(click to toggle)
def truncated_cmap(name, minval=0.3, maxval=0.9, n=6):
base = colormaps.get_cmap(name)
return lambda i: base(minval + (maxval - minval) * i / (n - 1))
mpl.rcdefaults()
mpl.rcParams.update(
{
"text.usetex": False,
"font.family": "serif",
"font.serif": ["DejaVu Serif"],
"axes.labelsize": 26,
"axes.titlesize": 24,
"legend.fontsize": 18,
"xtick.labelsize": 18,
"ytick.labelsize": 18,
}
)
ot.plot.setup.show_region_bounds = False…
(click to toggle)
ot.plot.setup.show_region_bounds = False
out_dir = Path(os.environ.get("OGS_TESTRUNNER_OUT_DIR", "_out"))
if not out_dir.exists():
out_dir.mkdir(parents=True)Great cell
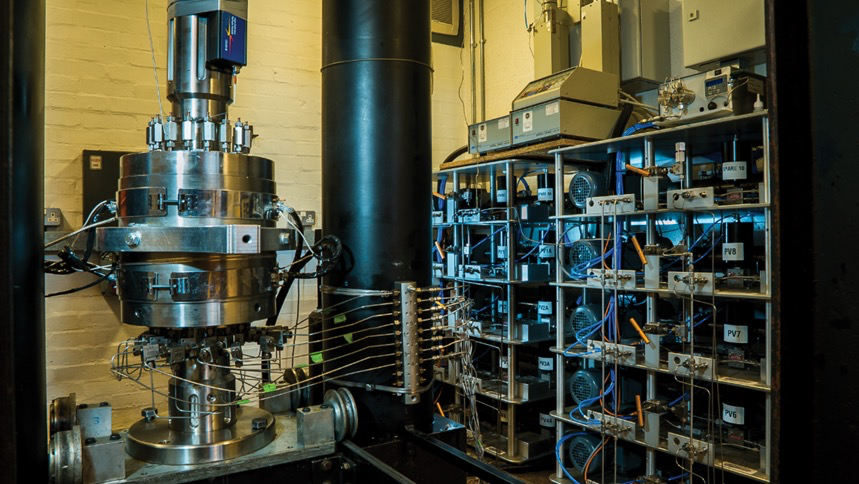
The GREAT cell is a poly-axial rock-testing device that reproduces subsurface conditions down to 3.5 km depth on 200 mm-diameter samples. It imposes a rotating stress field, injects fluid through a central borehole, and records both fiber-optic strain and pore-pressure data—providing a rich dataset for validating coupled hydro-mechanical models. For full details, see the GREAT cell benchmark docs: www.opengeosys.org/docs/benchmarks/small-deformations/greatcellm/
We distinguish two main types of benchmarks: intact and fractured rock tests. In the first type, an intact rock sample is subjected to rotating external stress conditions ($\texttt{M}_1$) and fluid is injected from a central borehole ($\texttt{HM}_1$):
- $\texttt{M}_1$: Intact rock sample under rotating boundary conditions.
- $\texttt{HM}_1$: Intact rock sample with fluid injection.
For the fractured rock samples, two subtypes are also considered: static and propagating fractures. Static fractures are studied as full ($\texttt{M}_{2a}$, $\texttt{HM}_{2a}$) and half-fractured ($\texttt{M}_{2b}$, $\texttt{HM}_{2b}$) versions to have a symmetric and non-symmetric case. Again, mechanical and hydro-mechanical versions are studied, the latter with fluid injection into the rock fracture:
- $\texttt{M}_{2a}$: Fully fractured sample.
- $\texttt{M}_{2b}$: Half-fractured sample.
- $\texttt{HM}_{2a}$: Fully fractured sample with inflow-outflow.
- $\texttt{HM}_{2b}$: Half-fractured sample with inflow-outflow.
To run this benchmark, you need to have OGS built with PETSc and PIP support. The following steps outline the process:
-
Configure & build OGS with PETSc & PIP
cmake -S ogs-source -B build-folder --preset release-petsc \ -DOGS_USE_PIP=ON -
Run the benchmark
cd build-folder/release-petsc ninja # OR make -j ctest -R nb-HMPhaseField/GreatCell -
Verify output files
ls build-folder/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCell
Material Properties
The material properties are provided in the following dictionary. The computational model incorporates two distinct elastic materials within its domain: a central circle ($r=0.097$ m) of rock surrounded by a rubber sheath in a 2D configuration.
materials = {…
(click to toggle)
materials = {
"Gneiss": {
"young_sample": 83.9e9, # Young's modulus (Pa)
"nu_sample": 0.21, # Poisson's ratio
"biot": 0.6, # Biot coefficient
"porosity": 0.001, # Porosity
"permeability": 1e-19, # Permeability (m²)
"density_solid": 2750, # Solid density (kg/m³)
"k_n": 200e9, # Normal stiffness (Pa/m)
"k_t": 100e9, # Tangential stiffness (Pa/m)
"c_f": 4.4e-10, # Fluid compressibility (Pa⁻¹)
"k_s": 4.82e10, # Solid bulk modulus (Pa)
"S_f": 4.4e-10, # Specific storage (Pa⁻¹)
"t_np": 10e6, # Peak normal traction (Pa)
"Gc": 50, # Fracture toughness (J/m²)
"w_init": 1e-6, # initial fracture width (m)
"fluid": {
"density": 1000.0, # Fluid density (kg/m³)
"viscosity": 1e-3, # Fluid viscosity (Pa·s)
"injectionFlowRate_Inlet": 4.167e-7, # Injection flow rate (m³/s)
"p_outlet": 3.45e6, # Outlet pressure (Pa)
},
"rubber_sheath": {
"young_modulus": 0.1e9, # Young's modulus (Pa)
"poisson_ratio": 0.4, # Poisson's ratio
"porosity": 0.001, # Porosity
"permeability": 1e-17, # Permeability (m²)
"density": 1500, # Density (kg/m³)
"biot": 0.0, # Biot coefficient
},
},
"Greywacke": {
"young_sample": 26.87e9, # Young's modulus (Pa)
"nu_sample": 0.27, # Poisson's ratio
"biot": 0.8, # Biot coefficient
"porosity": 0.005, # Porosity
"permeability": 2.58e-19, # Permeability (m²)
"density_solid": 2650, # Solid density (kg/m³)
"k_n": 100e9, # Normal stiffness (Pa/m)
"k_t": 50e9, # Tangential stiffness (Pa/m)
"c_f": 4.4e-10, # Fluid compressibility (Pa⁻¹)
"k_s": 1.95e10, # Solid bulk modulus (Pa)
"S_f": 4.4e-10, # Specific storage (Pa⁻¹)
"t_np": 10e6, # Peak normal traction (Pa)
"Gc": 30, # Fracture toughness (J/m²)
"w_init": 1e-6, # initial fracture width (m)
"fluid": {
"density": 1000.0, # Fluid density (kg/m³)
"viscosity": 1.0e-3, # Fluid viscosity (Pa·s)
"injectionFlowRate_Inlet": 4.167e-7, # Injection flow rate (m³/s)
"p_outlet": 3.45e6, # Outlet pressure (Pa)
},
"rubber_sheath": {
"young_modulus": 0.1e9, # Young's modulus (Pa)
"poisson_ratio": 0.4, # Poisson's ratio
"porosity": 0.001, # Porosity
"permeability": 1e-17, # Permeability (m²)
"density": 1500, # Density (kg/m³)
"biot": 0.0, # Biot coefficient
},
},
}
material_names = list(materials.keys())Loads
Table: Load Conditions
| Marker | PEE1 Angle to $\sigma_2$ | PEE1 & 1a | PEE2 & 2a | PEE3 & 3a | PEE4 & 4a | PEE5 & 5a | PEE6 & 6a | PEE7 & 7a | PEE8 & 8a |
|---|---|---|---|---|---|---|---|---|---|
| A | 0° | 10.0 | 6.64 | 4.46 | 1.17 | 1.0 | 3.82 | 7.80 | 9.95 |
| E | 22.5° | 9.95 | 10.0 | 6.64 | 4.46 | 1.17 | 1.0 | 3.82 | 7.80 |
| B | 45.0° | 7.80 | 9.95 | 10.0 | 6.64 | 4.46 | 1.17 | 1.0 | 3.82 |
| F | 67.5° | 3.82 | 7.80 | 9.95 | 10.0 | 6.64 | 4.46 | 1.17 | 1.0 |
| C | 90° | 1.0 | 3.82 | 7.80 | 9.95 | 10.0 | 6.64 | 4.46 | 1.17 |
All loads are in MPa. DSS loads are averages of adjacent PEEs.
Schematic loading conditions
loads = {…
(click to toggle)
loads = {
"A": [
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
7.80e6,
9.95e6,
],
"B": [
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
],
"C": [
1.0e6,
3.82e6,
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
],
}
angles_degrees = […
(click to toggle)
angles_degrees = [
0,
-22.5,
-45,
-67.5,
-90,
-112.5,
-135,
-157.5,
-180,
-202.5,
-225,
-247.5,
-270,
-292.5,
-315,
-337.5,
]
angles_radians = np.deg2rad(angles_degrees)
circle_radius = 6
circle_x = circle_radius * np.cos(angles_radians)
circle_y = circle_radius * np.sin(angles_radians)
fig, axs = plt.subplots(1, 3, figsize=(21, 7), facecolor="none")
for idx, (label, values) in enumerate(loads.items()):
ax = axs[idx]
ax.set_aspect("equal")
ax.axis("off")
formatted_values = [rf"${v / 1e6:.1f}$" for v in values]
scaled_values = [v / 2 for v in values]
circle = plt.Circle(
(0, 0),
circle_radius,
color="black",
fill=False,
linestyle="--",
linewidth=2,
)
ax.add_artist(circle)
top_points_x, top_points_y = [], []
for i, (x, y, value) in enumerate(
zip(circle_x, circle_y, scaled_values, strict=True)
):
unit_vector = np.array([x, y]) / circle_radius
line_end = np.array([x, y]) + unit_vector * value / 1e6
top_points_x.append(line_end[0])
top_points_y.append(line_end[1])
ax.annotate(
"",
xytext=line_end,
xy=(x, y),
arrowprops={
"arrowstyle": "-|>",
"color": "blue",
"lw": 2,
"mutation_scale": 15,
"fill": True,
},
)
angle_offset = 1.2 * unit_vector
angle_label = f"{-angles_degrees[i]}°"
ax.text(
x - angle_offset[0],
y - angle_offset[1],
angle_label,
fontsize=12,
ha="center",
va="center",
color="m",
)
value_offset = 0.5 * unit_vector
ax.text(
line_end[0] + 3.0 * value_offset[0],
line_end[1] + 1.8 * value_offset[1],
formatted_values[i],
fontsize=18,
ha="center",
weight="bold",
)
top_points_x.append(top_points_x[0])
top_points_y.append(top_points_y[0])
ax.plot(
top_points_x,
top_points_y,
color="green",
linestyle="-.",
lw=2,
marker="o",
markersize=6,
markerfacecolor="lightgreen",
markeredgewidth=1.0,
markeredgecolor="black",
)
ax.set_xlim([-12, 12])
ax.set_ylim([-12, 12])
ax.text(
0,
0,
f"Load {label}",
fontsize=32,
ha="center",
va="center",
family="serif",
)
plt.tight_layout()
output_path = Path(out_dir, "loads_A_B_C_schematic_with_angles.png")
plt.savefig(output_path, dpi=350, bbox_inches="tight", transparent=True)
plt.show()

Boundary Conditions
The boundary conditions applied in the simulation include both Dirichlet and Neumann conditions.
-
Dirichlet conditions (displacement control): \begin{equation*} \begin{cases} u_x(0, y) = 0, \quad u_y(0, y) = 0 & \quad \text{for } y = -0.09894 \text{ m}, \ u_y(x, 0) = 0, & \quad \text{for } x = -0.09894 \text{ m}. \end{cases} \end{equation*}
-
Neumann conditions: Normal stress $\sigma_{rr}$ is applied on PEEs and DSSs. The DSS stress is calculated as:
$$\sigma_\text{DSS}^i = \frac{\sigma_\text{PEE}^i + \sigma_\text{PEE}^{i+1}}{2}$$
Mesh generation of intact samples
Input
h = 0.005…
(click to toggle)
h = 0.005
meshname = "GreatCell"
mesh_path = Path(out_dir, "mesh_intact").resolve()
print(mesh_path)/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact
Boundary meshes
Gmsh
msh_file = mesh_GreatCell_intact(…
(click to toggle)
msh_file = mesh_GreatCell_intact(
lc=0.005,
lc2=h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path,
meshname=meshname,
mode="BC",
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 20%] Meshing curve 110 (Line)
Info : [ 20%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 30%] Meshing curve 118 (Line)
Info : [ 30%] Meshing curve 119 (Line)
Info : [ 30%] Meshing curve 120 (Line)
Info : [ 30%] Meshing curve 121 (Line)
Info : [ 30%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 40%] Meshing curve 126 (Line)
Info : [ 40%] Meshing curve 127 (Line)
Info : [ 40%] Meshing curve 128 (Line)
Info : [ 40%] Meshing curve 129 (Line)
Info : [ 40%] Meshing curve 130 (Line)
Info : [ 40%] Meshing curve 131 (Line)
Info : [ 40%] Meshing curve 132 (Line)
Info : [ 40%] Meshing curve 133 (Line)
Info : [ 50%] Meshing curve 134 (Line)
Info : [ 50%] Meshing curve 135 (Line)
Info : [ 50%] Meshing curve 136 (Line)
Info : [ 50%] Meshing curve 137 (Line)
Info : [ 50%] Meshing curve 138 (Line)
Info : [ 50%] Meshing curve 139 (Line)
Info : [ 50%] Meshing curve 140 (Line)
Info : [ 50%] Meshing curve 141 (Line)
Info : [ 60%] Meshing curve 142 (Line)
Info : [ 60%] Meshing curve 143 (Line)
Info : [ 60%] Meshing curve 144 (Line)
Info : [ 60%] Meshing curve 145 (Line)
Info : [ 60%] Meshing curve 146 (Line)
Info : [ 60%] Meshing curve 147 (Line)
Info : [ 60%] Meshing curve 148 (Line)
Info : [ 60%] Meshing curve 149 (Line)
Info : [ 70%] Meshing curve 150 (Line)
Info : [ 70%] Meshing curve 151 (Line)
Info : [ 70%] Meshing curve 152 (Line)
Info : [ 70%] Meshing curve 153 (Line)
Info : [ 70%] Meshing curve 154 (Line)
Info : [ 70%] Meshing curve 155 (Line)
Info : [ 70%] Meshing curve 156 (Line)
Info : [ 70%] Meshing curve 157 (Line)
Info : [ 80%] Meshing curve 158 (Line)
Info : [ 80%] Meshing curve 159 (Line)
Info : [ 80%] Meshing curve 160 (Line)
Info : [ 80%] Meshing curve 161 (Line)
Info : [ 80%] Meshing curve 162 (Line)
Info : [ 80%] Meshing curve 163 (Line)
Info : [ 80%] Meshing curve 164 (Line)
Info : [ 80%] Meshing curve 1234 (Line)
Info : [ 90%] Meshing curve 3005 (Circle)
Info : [ 90%] Meshing curve 3006 (Circle)
Info : [ 90%] Meshing curve 3007 (Circle)
Info : [ 90%] Meshing curve 3008 (Circle)
Info : [ 90%] Meshing curve 3015 (Circle)
Info : [ 90%] Meshing curve 3016 (Circle)
Info : [ 90%] Meshing curve 3017 (Circle)
Info : [ 90%] Meshing curve 3018 (Circle)
Info : [100%] Meshing curve 3025 (Circle)
Info : [100%] Meshing curve 3026 (Circle)
Info : [100%] Meshing curve 3027 (Circle)
Info : [100%] Meshing curve 3028 (Circle)
Info : [100%] Meshing curve 3035 (Circle)
Info : [100%] Meshing curve 3036 (Circle)
Info : [100%] Meshing curve 3037 (Circle)
Info : [100%] Meshing curve 3038 (Circle)
Info : Done meshing 1D (Wall 0.00355603s, CPU 0.004025s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 200 (Plane, Delaunay)
Info : [ 0%] Blossom: 2264 internal 88 closed
Info : [ 0%] Blossom recombination completed (Wall 0.0134373s, CPU 0.013416s): 740 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.776443, min Q = 0.460287
Info : [ 30%] Meshing surface 201 (Plane, Delaunay)
Info : [ 30%] Blossom: 2128 internal 208 closed
Info : [ 30%] Blossom recombination completed (Wall 0.0110471s, CPU 0.011035s): 744 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.715401, min Q = 0.462407
Info : [ 50%] Meshing surface 202 (Plane, Delaunay)
Info : [ 50%] Blossom: 600 internal 240 closed
Info : [ 50%] Blossom recombination completed (Wall 0.0018256s, CPU 0.001844s): 120 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.983333, min Q = 0.983333
Info : [ 70%] Meshing surface 203 (Plane, Delaunay)
Info : [ 70%] Blossom: 344 internal 248 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00156871s, CPU 0.001582s): 132 quads, 16 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.59063, min Q = 0.274456
Info : [ 90%] Meshing surface 204 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.0564905s, CPU 0.056597s)
Info : 2097 nodes 3002 elements
Info : Writing '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/GreatCell.msh'
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/domain.vtu (160 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_Inlet.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_p_right.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_p_top.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_p_left.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_p_bottom.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE1.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE2.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE3.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE4.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE5.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE6.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE7.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE8.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE1a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE2a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE3a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE4a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE5a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE6a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE7a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE8a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS1.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS2.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS3.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS4.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS5.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS6.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS7.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS8.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS1a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS2a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS3a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS4a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS5a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS6a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS7a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS8a.vtu (4 cells)
[0m[33m2025-10-23 14:32:15.655 ( 1.995s) [ 7FD7BBBB7BC0]vtkXOpenGLRenderWindow.:1416 WARN| bad X server connection. DISPLAY=[0m

Convert .msh to an OGS-compatible mesh
msh_path = Path(mesh_path, f"{meshname}.msh")…
(click to toggle)
msh_path = Path(mesh_path, f"{meshname}.msh")
meshes = ot.meshes_from_gmsh(filename=msh_path, dim=[1], reindex=True, log=False)
for name, mesh in meshes.items():
print(f"{name}: {mesh.n_cells} cells")
pv.save_meshio(Path(mesh_path, f"{name}.vtu"), mesh)domain: 160 cells
physical_group_Inlet: 1 cells
physical_group_p_right: 1 cells
physical_group_p_top: 1 cells
physical_group_p_left: 1 cells
physical_group_p_bottom: 1 cells
physical_group_PEE1: 6 cells
physical_group_PEE2: 6 cells
physical_group_PEE3: 6 cells
physical_group_PEE4: 6 cells
physical_group_PEE5: 6 cells
physical_group_PEE6: 6 cells
physical_group_PEE7: 6 cells
physical_group_PEE8: 6 cells
physical_group_PEE1a: 6 cells
physical_group_PEE2a: 6 cells
physical_group_PEE3a: 6 cells
physical_group_PEE4a: 6 cells
physical_group_PEE5a: 6 cells
physical_group_PEE6a: 6 cells
physical_group_PEE7a: 6 cells
physical_group_PEE8a: 6 cells
physical_group_DSS1: 4 cells
physical_group_DSS2: 4 cells
physical_group_DSS3: 4 cells
physical_group_DSS4: 4 cells
physical_group_DSS5: 4 cells
physical_group_DSS6: 4 cells
physical_group_DSS7: 4 cells
physical_group_DSS8: 4 cells
physical_group_DSS1a: 4 cells
physical_group_DSS2a: 4 cells
physical_group_DSS3a: 4 cells
physical_group_DSS4a: 4 cells
physical_group_DSS5a: 4 cells
physical_group_DSS6a: 4 cells
physical_group_DSS7a: 4 cells
physical_group_DSS8a: 4 cells
Visualization of boundary meshes
plotter = pv.Plotter()…
(click to toggle)
plotter = pv.Plotter()
for name, mesh in meshes.items():
if mesh.active_scalars is not None:
plotter.add_mesh(
mesh,
scalars=mesh.active_scalars_name,
cmap="tab20",
show_edges=False,
opacity=0.7,
)
else:
plotter.add_mesh(mesh, show_edges=False, opacity=0.7, label=name)
clean_name = name.replace("physical_group_", "")
center = mesh.center
direction = np.array(center) - np.array([0, 0, 0])
direction[:2] = direction[:2] / (np.linalg.norm(direction[:2]) + 1e-8)
offset = center + 0.025 * direction
plotter.add_point_labels(
[offset], [clean_name], font_size=12, point_size=0, text_color="black"
)
plotter.view_xy()
plotter.enable_parallel_projection()
plotter.show()

Computational domain mesh
Gmsh
msh_file = mesh_GreatCell_intact(…
(click to toggle)
msh_file = mesh_GreatCell_intact(
lc=0.005,
lc2=h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path,
meshname=meshname,
mode="domain",
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 20%] Meshing curve 110 (Line)
Info : [ 20%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 30%] Meshing curve 118 (Line)
Info : [ 30%] Meshing curve 119 (Line)
Info : [ 30%] Meshing curve 120 (Line)
Info : [ 30%] Meshing curve 121 (Line)
Info : [ 30%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 40%] Meshing curve 126 (Line)
Info : [ 40%] Meshing curve 127 (Line)
Info : [ 40%] Meshing curve 128 (Line)
Info : [ 40%] Meshing curve 129 (Line)
Info : [ 40%] Meshing curve 130 (Line)
Info : [ 40%] Meshing curve 131 (Line)
Info : [ 40%] Meshing curve 132 (Line)
Info : [ 40%] Meshing curve 133 (Line)
Info : [ 50%] Meshing curve 134 (Line)
Info : [ 50%] Meshing curve 135 (Line)
Info : [ 50%] Meshing curve 136 (Line)
Info : [ 50%] Meshing curve 137 (Line)
Info : [ 50%] Meshing curve 138 (Line)
Info : [ 50%] Meshing curve 139 (Line)
Info : [ 50%] Meshing curve 140 (Line)
Info : [ 50%] Meshing curve 141 (Line)
Info : [ 60%] Meshing curve 142 (Line)
Info : [ 60%] Meshing curve 143 (Line)
Info : [ 60%] Meshing curve 144 (Line)
Info : [ 60%] Meshing curve 145 (Line)
Info : [ 60%] Meshing curve 146 (Line)
Info : [ 60%] Meshing curve 147 (Line)
Info : [ 60%] Meshing curve 148 (Line)
Info : [ 60%] Meshing curve 149 (Line)
Info : [ 70%] Meshing curve 150 (Line)
Info : [ 70%] Meshing curve 151 (Line)
Info : [ 70%] Meshing curve 152 (Line)
Info : [ 70%] Meshing curve 153 (Line)
Info : [ 70%] Meshing curve 154 (Line)
Info : [ 70%] Meshing curve 155 (Line)
Info : [ 70%] Meshing curve 156 (Line)
Info : [ 70%] Meshing curve 157 (Line)
Info : [ 80%] Meshing curve 158 (Line)
Info : [ 80%] Meshing curve 159 (Line)
Info : [ 80%] Meshing curve 160 (Line)
Info : [ 80%] Meshing curve 161 (Line)
Info : [ 80%] Meshing curve 162 (Line)
Info : [ 80%] Meshing curve 163 (Line)
Info : [ 80%] Meshing curve 164 (Line)
Info : [ 80%] Meshing curve 1234 (Line)
Info : [ 90%] Meshing curve 3005 (Circle)
Info : [ 90%] Meshing curve 3006 (Circle)
Info : [ 90%] Meshing curve 3007 (Circle)
Info : [ 90%] Meshing curve 3008 (Circle)
Info : [ 90%] Meshing curve 3015 (Circle)
Info : [ 90%] Meshing curve 3016 (Circle)
Info : [ 90%] Meshing curve 3017 (Circle)
Info : [ 90%] Meshing curve 3018 (Circle)
Info : [100%] Meshing curve 3025 (Circle)
Info : [100%] Meshing curve 3026 (Circle)
Info : [100%] Meshing curve 3027 (Circle)
Info : [100%] Meshing curve 3028 (Circle)
Info : [100%] Meshing curve 3035 (Circle)
Info : [100%] Meshing curve 3036 (Circle)
Info : [100%] Meshing curve 3037 (Circle)
Info : [100%] Meshing curve 3038 (Circle)
Info : Done meshing 1D (Wall 0.00344029s, CPU 0.005011s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 200 (Plane, Delaunay)
Info : [ 0%] Blossom: 2264 internal 88 closed
Info : [ 0%] Blossom recombination completed (Wall 0.0128515s, CPU 0.012836s): 740 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.776443, min Q = 0.460287
Info : [ 30%] Meshing surface 201 (Plane, Delaunay)
Info : [ 30%] Blossom: 2128 internal 208 closed
Info : [ 30%] Blossom recombination completed (Wall 0.0107631s, CPU 0.010765s): 744 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.715401, min Q = 0.462407
Info : [ 50%] Meshing surface 202 (Plane, Delaunay)
Info : [ 50%] Blossom: 600 internal 240 closed
Info : [ 50%] Blossom recombination completed (Wall 0.00178104s, CPU 0.001793s): 120 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.983333, min Q = 0.983333
Info : [ 70%] Meshing surface 203 (Plane, Delaunay)
Info : [ 70%] Blossom: 344 internal 248 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00151944s, CPU 0.001538s): 132 quads, 16 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.59063, min Q = 0.274456
Info : [ 90%] Meshing surface 204 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.0546337s, CPU 0.054751s)
Info : 2097 nodes 3002 elements
Info : Writing '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/GreatCell.msh'
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/domain.vtu (2296 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_Central_sample.vtu (740 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_OuterPart_sample.vtu (1012 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_Rubber_sheath.vtu (544 cells)

Convert .msh to an OGS-compatible mesh
msh_path = Path(mesh_path, f"{meshname}.msh")…
(click to toggle)
msh_path = Path(mesh_path, f"{meshname}.msh")
meshes_volume = ot.meshes_from_gmsh(
filename=msh_path, dim=[1, 2], reindex=True, log=False
)
for name, mesh in meshes_volume.items():
print(f"{name}: {mesh.n_cells} cells")
pv.save_meshio(Path(mesh_path, f"{name}.vtu"), mesh)domain: 2296 cells
physical_group_Central_sample: 740 cells
physical_group_OuterPart_sample: 1012 cells
physical_group_Rubber_sheath: 544 cells
Visualization of computational domain mesh
plotter = pv.Plotter()…
(click to toggle)
plotter = pv.Plotter()
for name, mesh in meshes_volume.items():
if mesh.active_scalars is not None:
plotter.add_mesh(
mesh,
scalars=mesh.active_scalars_name,
cmap="Set1",
show_edges=False,
opacity=0.7,
)
else:
plotter.add_mesh(mesh, show_edges=False, opacity=0.7, label=name)
clean_name = name.replace("physical_group_", "")
center = mesh.center
direction = np.array(center) - np.array([0, 0, 0])
direction[:2] = direction[:2] / (np.linalg.norm(direction[:2]) + 1e-8)
offset = center + 0.025 * direction
plotter.add_point_labels(
[offset], [clean_name], font_size=12, point_size=0, text_color="black"
)
plotter.view_xy()
plotter.enable_parallel_projection()
plotter.show()

mesh_dir = Path(mesh_path).resolve()…
(click to toggle)
mesh_dir = Path(mesh_path).resolve()
run(
[
"NodeReordering",
"-i",
str(mesh_dir.joinpath("domain.vtu")),
"-o",
str(mesh_dir.joinpath("domain.vtu")),
],
cwd=mesh_dir,
check=True,
)
physical_groups = [
"physical_group_DSS1.vtu",
"physical_group_DSS1a.vtu",
"physical_group_DSS2.vtu",
"physical_group_DSS2a.vtu",
"physical_group_DSS3.vtu",
"physical_group_DSS3a.vtu",
"physical_group_DSS4.vtu",
"physical_group_DSS4a.vtu",
"physical_group_DSS5.vtu",
"physical_group_DSS5a.vtu",
"physical_group_DSS6.vtu",
"physical_group_DSS6a.vtu",
"physical_group_DSS7.vtu",
"physical_group_DSS7a.vtu",
"physical_group_DSS8.vtu",
"physical_group_DSS8a.vtu",
"physical_group_PEE1.vtu",
"physical_group_PEE1a.vtu",
"physical_group_PEE2.vtu",
"physical_group_PEE2a.vtu",
"physical_group_PEE3.vtu",
"physical_group_PEE3a.vtu",
"physical_group_PEE4.vtu",
"physical_group_PEE4a.vtu",
"physical_group_PEE5.vtu",
"physical_group_PEE5a.vtu",
"physical_group_PEE6.vtu",
"physical_group_PEE6a.vtu",
"physical_group_PEE7.vtu",
"physical_group_PEE7a.vtu",
"physical_group_PEE8.vtu",
"physical_group_PEE8a.vtu",
"physical_group_p_bottom.vtu",
"physical_group_p_left.vtu",
"physical_group_p_right.vtu",
"physical_group_p_top.vtu",
"physical_group_Inlet.vtu",
]
group_paths = [str(mesh_dir.joinpath(name)) for name in physical_groups]
run(
[
"identifySubdomains",
"-f",
"-m",
str(mesh_dir.joinpath("domain.vtu")),
"--",
*group_paths,
],
cwd=mesh_dir,
check=True,
)[0] info: Reordering nodes...
[0] info: Method: Reversing order of nodes unless it is considered correct by the OGS6 standard, i.e. such that det(J) > 0, where J is the Jacobian of the global-to-local coordinate transformation.
[0] info: Corrected 0 elements.
[0] info: VTU file written.
[0] info: Mesh reading time: 0.0153237 s
[0] info: MeshNodeSearcher construction time: 0.000126852 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.0155e-05 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000155375 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.242e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000113111 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.91e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000110016 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.91e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000109846 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.71e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000120342 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.21e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000112019 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.12e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.0001129 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.62e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000111078 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.71e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000111018 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.62e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000110267 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.81e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000109445 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.11e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000109756 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.01e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000116506 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.22e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000111118 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.21e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.0001124 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.062e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000111849 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.312e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000110937 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.131e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000112691 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.161e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000113863 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.171e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.00011281 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.22e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000117588 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.092e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000114453 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.81e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000112981 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.172e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000113692 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.141e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000116456 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.192e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000114693 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.082e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000117287 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.152e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000117367 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.342e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000121774 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.072e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000115374 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.022e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000113662 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.92e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000113992 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.61e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_bottom' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000113521 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.31e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_left' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000113712 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.3e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_right' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000113942 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.41e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_top' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000114092 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.9e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_Inlet' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000117828 s
[0] info: identifySubdomains time: 0.00460522 s
[0] info: writing time: 0.0357162 s
[0] info: Entire run time: -2.26768e+06 s
CompletedProcess(args=['identifySubdomains', '-f', '-m', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/domain.vtu', '--', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS1.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS1a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS2.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS2a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS3.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS3a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS4.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS4a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS5.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS5a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS6.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS6a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS7.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS7a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS8.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_DSS8a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE1.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE1a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE2.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE2a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE3.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE3a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE4.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE4a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE5.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE5a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE6.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE6a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE7.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE7a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE8.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_PEE8a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_p_bottom.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_p_left.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_p_right.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_p_top.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact/physical_group_Inlet.vtu'], returncode=0)
Intact Samples ($\texttt{M}_1$)
The primary objective of the first benchmark exercise is to evaluate the mechanical deformation of different rock samples in the GREAT cell under various boundary conditions in a 2D plane strain setup.
This benchmark considers a plane strain triaxial stress condition, where the in-plane principal stresses are defined as:
$$ \sigma_2 = 10\ \text{MPa},\quad \sigma_3 = 1\ \text{MPa} $$The out-of-plane stress component, $\sigma_1$, is governed by the plane strain constraint:
$$ \sigma_1 = \nu (\sigma_2 + \sigma_3) $$This loading condition is applied to both Greywacke and Gneiss samples
Run the simulation
Inputs
# Times for load curves…
(click to toggle)
# Times for load curves
times = "0.0 1000. 3500"
simulation_end_time = 3500.0
n_fracture_p_ncs = 0
model_type = "M1"
output_prefix = "M1_VPF"
# Load
PEE_load_values = {
"A": [10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6, 9.95e6],
"B": [7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6],
"C": [1.0e6, 3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6],
}
prj_file = Path("M1_VPF.prj")
prj = ot.Project(input_file=prj_file, output_file=Path(out_dir, f"{output_prefix}.prj"))Run OGS
# Create SingleOGSModel…
(click to toggle)
# Create SingleOGSModel
sing_ogs_model = SingleOGSModel(
model=prj,
out_dir=out_dir,
mesh_path=mesh_path,
output_prefix=output_prefix,
method="VPF",
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
materials=materials,
)
# Run simulations
vtu_files_dict = sing_ogs_model.run_simulations_with_fracture(
times=times,
base_project_file=prj_file,
mesh_path=mesh_path,
load_cases=PEE_load_values,
material_names=material_names,
materials=materials,
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
output_prefix=output_prefix,
out_dir=out_dir,
method="VPF",
fracture_model_type="VolumetricDeviatoric",
mesh_size=h,
)mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M1_VPF.prj
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M1_VPF_A.prj
============================================================
Running simulation for load case: A with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.01 based on mesh size 0.005
ls 0.01
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M1_VPF_A.prj
Status: finished successfully.
Execution took 8.75036907196045 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.01 based on mesh size 0.005
ls 0.01
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M1_VPF_A.prj
Status: finished successfully.
Execution took 9.211067199707031 s
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M1_VPF_B.prj
============================================================
Running simulation for load case: B with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.01 based on mesh size 0.005
ls 0.01
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M1_VPF_B.prj
Status: finished successfully.
Execution took 8.643445253372192 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.01 based on mesh size 0.005
ls 0.01
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M1_VPF_B.prj
Status: finished successfully.
Execution took 8.797649621963501 s
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M1_VPF_C.prj
============================================================
Running simulation for load case: C with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.01 based on mesh size 0.005
ls 0.01
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M1_VPF_C.prj
Status: finished successfully.
Execution took 8.551372528076172 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.01 based on mesh size 0.005
ls 0.01
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M1_VPF_C.prj
Status: finished successfully.
Execution took 8.822606325149536 s
Post-processing
Volumetric strain vs angle at probe circle
json_path = Path("./external_data_dict.py").resolve()…
(click to toggle)
json_path = Path("./external_data_dict.py").resolve()
print(f"[DEBUG] Trying path: {json_path}")
if json_path.exists():
from external_data_dict import external_data
else:
print("[WARNING] External data dict not found! Skipping...")
external_data = None
plotter = Plotter(
output_dir=out_dir,
save_extracted_data=True,
)
data_dir = Path("external_data")
external_data = Plotter.load_external_data(data_dir, benchmark_tag="M1")
plotter.plot_volumetric_strain_vs_angle(
vtu_files_dict,
model_type="M1",
ylim_range=[-7.5, 2.5],
layout="subplots",
external_data=external_data["strain"],
)[DEBUG] Trying path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell/external_data_dict.py
[WARNING] External data dict not found! Skipping...

Profiles
plotter.plot_field_variables(vtu_files_dict)Load: A, File 0, Benchmark: M1, Material: Gneiss

Load: A, File 1, Benchmark: M1, Material: Greywacke

Load: B, File 0, Benchmark: M1, Material: Gneiss

Load: B, File 1, Benchmark: M1, Material: Greywacke

Load: C, File 0, Benchmark: M1, Material: Gneiss

Load: C, File 1, Benchmark: M1, Material: Greywacke

Intact rock sample with fluid injection ($\texttt{HM}_1$)
This benchmark, which does not consider any fracture, is designed to verify the basic computational setting for hydro-mechanical simulations. In addition to the mechanical loads, a zero constant pore pressure is prescribed at the outer boundary. At the center of sample, fluid is injected at a rate of $Q_0^{\text{v}} = 2.085 \times 10^{-9}$ m$^3$/s
The hydro-mechanical simulations follows a two-stage process: a 3000s equilibrium phase under mechanical loading to stabilize initial conditions, followed by a 500s fluid injection phase to model fluid flow. This loading condition is applied to both Greywacke and Gneiss samples
Run the simulation
Inputs
# Times for load curves…
(click to toggle)
# Times for load curves
times = "0.0 1000. 3500"
simulation_end_time = 3500.0
n_fracture_p_ncs = 0
model_type = "HM1"
output_prefix = "HM1_VPF"
# Load
PEE_load_values = {
"A": [10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6, 9.95e6],
"B": [7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6],
"C": [1.0e6, 3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6],
}
for p in materials.values():
p["fluid"][
"injectionFlowRate_Inlet"
] = 2.085e-6 # Injection flow rate m³/s (125 ml/min)
prj_file = Path("HM1_VPF.prj")
prj = ot.Project(input_file=prj_file, output_file=Path(out_dir, f"{output_prefix}.prj"))Run OGS
# Create SingleOGSModel…
(click to toggle)
# Create SingleOGSModel
sing_ogs_model = SingleOGSModel(
model=prj,
out_dir=out_dir,
mesh_path=mesh_path,
output_prefix=output_prefix,
method="VPF",
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
materials=materials,
)
# Run simulations
vtu_files_dict_HM = sing_ogs_model.run_simulations_with_fracture(
times=times,
base_project_file=prj_file,
mesh_path=mesh_path,
load_cases=PEE_load_values,
material_names=material_names,
materials=materials,
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
output_prefix=output_prefix,
out_dir=out_dir,
method="VPF",
fracture_model_type="VolumetricDeviatoric",
mesh_size=h,
)mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM1_VPF.prj
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM1_VPF_A.prj
============================================================
Running simulation for load case: A with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.01 based on mesh size 0.005
ls 0.01
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM1_VPF_A.prj
Status: finished successfully.
Execution took 22.43727159500122 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.01 based on mesh size 0.005
ls 0.01
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM1_VPF_A.prj
Status: finished successfully.
Execution took 30.30357265472412 s
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM1_VPF_B.prj
============================================================
Running simulation for load case: B with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.01 based on mesh size 0.005
ls 0.01
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM1_VPF_B.prj
Status: finished successfully.
Execution took 22.425955533981323 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.01 based on mesh size 0.005
ls 0.01
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM1_VPF_B.prj
Status: finished successfully.
Execution took 30.385088205337524 s
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM1_VPF_C.prj
============================================================
Running simulation for load case: C with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.01 based on mesh size 0.005
ls 0.01
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM1_VPF_C.prj
Status: finished successfully.
Execution took 22.32820153236389 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.01 based on mesh size 0.005
ls 0.01
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM1_VPF_C.prj
Status: finished successfully.
Execution took 29.625319242477417 s
Post-processing
Volumetric strain vs angle at probe circle
json_path = Path("./external_data_dict.py").resolve()…
(click to toggle)
json_path = Path("./external_data_dict.py").resolve()
print(f"[DEBUG] Trying path: {json_path}")
if json_path.exists():
from external_data_dict import external_data
else:
print("[WARNING] External data dict not found! Skipping...")
external_data = None
data_dir = Path("external_data")
external_data = Plotter.load_external_data(data_dir, benchmark_tag="HM1")
plotter = Plotter(
output_dir=out_dir,
save_extracted_data=True,
)
plotter.plot_volumetric_strain_vs_angle(
vtu_files_dict_HM,
model_type="HM1",
ylim_range=[-7.5, 2.5],
layout="subplots",
external_data=external_data["strain"],
)[DEBUG] Trying path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell/external_data_dict.py
[WARNING] External data dict not found! Skipping...

Profiles
plotter.plot_field_variables(vtu_files_dict_HM)Load: A, File 0, Benchmark: HM1, Material: Gneiss

Load: A, File 1, Benchmark: HM1, Material: Greywacke

Load: B, File 0, Benchmark: HM1, Material: Gneiss

Load: B, File 1, Benchmark: HM1, Material: Greywacke

Load: C, File 0, Benchmark: HM1, Material: Gneiss

Load: C, File 1, Benchmark: HM1, Material: Greywacke

Mesh generation of fractured samples
Input
h = 0.001…
(click to toggle)
h = 0.001
meshname = "GreatCell"
mesh_path_VPF = Path(out_dir, "mesh_GreatCell_VPF").resolve()Boundary meshes
Gmsh
msh_file_VPF = mesh_GreatCell_VPF(…
(click to toggle)
msh_file_VPF = mesh_GreatCell_VPF(
lc2=h,
lc=20 * h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path_VPF,
meshname=meshname,
mode="BC",
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 10%] Meshing curve 110 (Line)
Info : [ 10%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 20%] Meshing curve 118 (Line)
Info : [ 20%] Meshing curve 119 (Line)
Info : [ 20%] Meshing curve 120 (Line)
Info : [ 20%] Meshing curve 121 (Line)
Info : [ 20%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 30%] Meshing curve 126 (Line)
Info : [ 30%] Meshing curve 127 (Line)
Info : [ 30%] Meshing curve 128 (Line)
Info : [ 30%] Meshing curve 129 (Line)
Info : [ 30%] Meshing curve 130 (Line)
Info : [ 30%] Meshing curve 131 (Line)
Info : [ 30%] Meshing curve 132 (Line)
Info : [ 30%] Meshing curve 133 (Line)
Info : [ 40%] Meshing curve 134 (Line)
Info : [ 40%] Meshing curve 135 (Line)
Info : [ 40%] Meshing curve 136 (Line)
Info : [ 40%] Meshing curve 137 (Line)
Info : [ 40%] Meshing curve 138 (Line)
Info : [ 40%] Meshing curve 139 (Line)
Info : [ 40%] Meshing curve 140 (Line)
Info : [ 40%] Meshing curve 141 (Line)
Info : [ 40%] Meshing curve 142 (Line)
Info : [ 40%] Meshing curve 143 (Line)
Info : [ 40%] Meshing curve 144 (Line)
Info : [ 50%] Meshing curve 145 (Line)
Info : [ 50%] Meshing curve 146 (Line)
Info : [ 50%] Meshing curve 147 (Line)
Info : [ 50%] Meshing curve 148 (Line)
Info : [ 50%] Meshing curve 149 (Line)
Info : [ 50%] Meshing curve 150 (Line)
Info : [ 50%] Meshing curve 151 (Line)
Info : [ 50%] Meshing curve 152 (Line)
Info : [ 50%] Meshing curve 153 (Line)
Info : [ 50%] Meshing curve 154 (Line)
Info : [ 60%] Meshing curve 155 (Line)
Info : [ 60%] Meshing curve 156 (Line)
Info : [ 60%] Meshing curve 157 (Line)
Info : [ 60%] Meshing curve 158 (Line)
Info : [ 60%] Meshing curve 159 (Line)
Info : [ 60%] Meshing curve 160 (Line)
Info : [ 60%] Meshing curve 161 (Line)
Info : [ 60%] Meshing curve 162 (Line)
Info : [ 60%] Meshing curve 163 (Line)
Info : [ 60%] Meshing curve 164 (Line)
Info : [ 60%] Meshing curve 401 (Circle)
Info : [ 70%] Meshing curve 402 (Circle)
Info : [ 70%] Meshing curve 403 (Circle)
Info : [ 70%] Meshing curve 404 (Circle)
Info : [ 70%] Meshing curve 411 (Circle)
Info : [ 70%] Meshing curve 412 (Circle)
Info : [ 70%] Meshing curve 413 (Circle)
Info : [ 70%] Meshing curve 414 (Circle)
Info : [ 70%] Meshing curve 421 (Circle)
Info : [ 70%] Meshing curve 422 (Circle)
Info : [ 70%] Meshing curve 423 (Circle)
Info : [ 70%] Meshing curve 424 (Circle)
Info : [ 80%] Meshing curve 431 (Circle)
Info : [ 80%] Meshing curve 432 (Circle)
Info : [ 80%] Meshing curve 433 (Circle)
Info : [ 80%] Meshing curve 434 (Circle)
Info : [ 80%] Meshing curve 501 (Line)
Info : [ 80%] Meshing curve 502 (Line)
Info : [ 80%] Meshing curve 503 (Line)
Info : [ 80%] Meshing curve 504 (Line)
Info : [ 80%] Meshing curve 505 (Line)
Info : [ 80%] Meshing curve 506 (Line)
Info : [ 80%] Meshing curve 507 (Line)
Info : [ 90%] Meshing curve 508 (Line)
Info : [ 90%] Meshing curve 509 (Line)
Info : [ 90%] Meshing curve 510 (Line)
Info : [ 90%] Meshing curve 511 (Line)
Info : [ 90%] Meshing curve 512 (Line)
Info : [ 90%] Meshing curve 513 (Line)
Info : [ 90%] Meshing curve 514 (Line)
Info : [ 90%] Meshing curve 515 (Line)
Info : [ 90%] Meshing curve 516 (Line)
Info : [ 90%] Meshing curve 517 (Line)
Info : [ 90%] Meshing curve 518 (Line)
Info : [100%] Meshing curve 519 (Line)
Info : [100%] Meshing curve 520 (Line)
Info : [100%] Meshing curve 521 (Line)
Info : [100%] Meshing curve 522 (Line)
Info : [100%] Meshing curve 523 (Line)
Info : [100%] Meshing curve 524 (Line)
Info : [100%] Meshing curve 525 (Line)
Info : [100%] Meshing curve 526 (Line)
Info : [100%] Meshing curve 5040 (Line)
Info : [100%] Meshing curve 5120 (Line)
Info : Done meshing 1D (Wall 0.0287631s, CPU 0.030068s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 701 (Plane, Frontal-Delaunay for Quads)
Info : [ 0%] Blossom: 51 internal 52 closed
Info : [ 0%] Blossom recombination completed (Wall 0.000111449s, CPU 0.000134s): 26 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 10%] Meshing surface 702 (Plane, Frontal-Delaunay for Quads)
Info : [ 10%] Blossom: 79 internal 80 closed
Info : [ 10%] Blossom recombination completed (Wall 0.000160072s, CPU 0.000185s): 40 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 20%] Meshing surface 703 (Plane, Frontal-Delaunay for Quads)
Info : [ 20%] Blossom: 129 internal 130 closed
Info : [ 20%] Blossom recombination completed (Wall 0.000266273s, CPU 0.000285s): 65 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 20%] Meshing surface 704 (Plane, Frontal-Delaunay for Quads)
Info : [ 20%] Blossom recombination completed (Wall 0.00741524s, CPU 0.007701s): 639 quads, 133 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.843335, min Q = 0.363603
Info : [ 30%] Meshing surface 705 (Plane, Frontal-Delaunay for Quads)
Info : [ 30%] Blossom recombination completed (Wall 0.00575167s, CPU 0.005797s): 665 quads, 125 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.844908, min Q = 0.460748
Info : [ 30%] Meshing surface 706 (Plane, Frontal-Delaunay for Quads)
Info : [ 30%] Blossom: 49 internal 50 closed
Info : [ 30%] Blossom recombination completed (Wall 0.000148424s, CPU 0.00017s): 25 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 40%] Meshing surface 707 (Plane, Frontal-Delaunay for Quads)
Info : [ 40%] Blossom: 49 internal 50 closed
Info : [ 40%] Blossom recombination completed (Wall 0.000148134s, CPU 0.000163s): 25 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 40%] Meshing surface 708 (Plane, Frontal-Delaunay for Quads)
Info : [ 40%] Blossom: 1190 internal 132 closed
Info : [ 40%] Blossom recombination completed (Wall 0.00789148s, CPU 0.007888s): 416 quads, 0 triangles, 0 invalid quads, 1 quads with Q < 0.1, avg Q = 0.744994, min Q = 0.0703248
Info : [ 50%] Meshing surface 709 (Plane, Frontal-Delaunay for Quads)
Info : [ 50%] Blossom: 1130 internal 132 closed
Info : [ 50%] Blossom recombination completed (Wall 0.00755608s, CPU 0.007551s): 395 quads, 0 triangles, 0 invalid quads, 3 quads with Q < 0.1, avg Q = 0.762586, min Q = 0.0293523
Info : [ 60%] Meshing surface 710 (Plane, Frontal-Delaunay for Quads)
Info : [ 60%] Blossom: 7 internal 8 closed
Info : [ 60%] Blossom recombination completed (Wall 0.000101995s, CPU 0.000122s): 4 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 60%] Meshing surface 711 (Plane, Frontal-Delaunay for Quads)
Info : [ 60%] Blossom: 7 internal 8 closed
Info : [ 60%] Blossom recombination completed (Wall 8.60109e-05s, CPU 0.000102s): 4 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 70%] Meshing surface 712 (Plane, Frontal-Delaunay for Quads)
Info : [ 70%] Blossom: 197 internal 103 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00105376s, CPU 0.001078s): 80 quads, 6 triangles, 0 invalid quads, 6 quads with Q < 0.1, avg Q = 0.564073, min Q = 0.0244822
Info : [ 70%] Meshing surface 713 (Plane, Frontal-Delaunay for Quads)
Info : [ 70%] Blossom: 191 internal 102 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00102402s, CPU 0.001044s): 75 quads, 12 triangles, 0 invalid quads, 8 quads with Q < 0.1, avg Q = 0.52015, min Q = 0.0178184
Info : [ 80%] Meshing surface 714 (Plane, Frontal-Delaunay for Quads)
Info : [ 80%] Blossom: 5 internal 6 closed
Info : [ 80%] Blossom recombination completed (Wall 9.56547e-05s, CPU 0.000116s): 3 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 80%] Meshing surface 715 (Plane, Frontal-Delaunay for Quads)
Info : [ 80%] Blossom: 5 internal 6 closed
Info : [ 80%] Blossom recombination completed (Wall 8.52891e-05s, CPU 0.000101s): 3 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 90%] Meshing surface 716 (Plane, Frontal-Delaunay for Quads)
Info : [ 90%] Blossom: 67 internal 68 closed
Info : [ 90%] Blossom recombination completed (Wall 0.000246193s, CPU 0.00026s): 18 quads, 32 triangles, 0 invalid quads, 4 quads with Q < 0.1, avg Q = 0.275269, min Q = 0.0598336
Info : [ 90%] Meshing surface 717 (Plane, Frontal-Delaunay for Quads)
Info : [ 90%] Blossom: 67 internal 68 closed
Info : [ 90%] Blossom recombination completed (Wall 0.00024459s, CPU 0.000261s): 18 quads, 32 triangles, 0 invalid quads, 4 quads with Q < 0.1, avg Q = 0.275269, min Q = 0.0598336
Info : [100%] Meshing surface 718 (Plane, Frontal-Delaunay for Quads)
Info : [100%] Blossom: 130 internal 130 closed
Info : [100%] Blossom recombination completed (Wall 0.000422479s, CPU 0.000443s): 50 quads, 30 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.144988, min Q = 0.125
Info : Done meshing 2D (Wall 0.0791789s, CPU 0.080171s)
Info : 2786 nodes 3806 elements
Info : Writing '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/GreatCell.msh'
Warning : Cannot apply Blossom: odd number of triangles (1413) in surface 704
Warning : Cannot apply Blossom: odd number of triangles (1455) in surface 705
Warning : ------------------------------
Warning : Mesh generation error summary
Warning : 2 warnings
Warning : 0 errors
Warning : Check the full log for details
Warning : ------------------------------
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/domain.vtu (100 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_right.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_top.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_left.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_bottom.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE1.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE2.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE3.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE4.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE5.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE6.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE7.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE8.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE1a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE2a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE3a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE4a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE5a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE6a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE7a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE8a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS1.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS2.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS3.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS4.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS5.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS6.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS7.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS8.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS1a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS2a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS3a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS4a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS5a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS6a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS7a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS8a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Inlet.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_R_embeddedFracture.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_R_fullFracture.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_L_fullFracture.vtu (1 cells)

Convert .msh to an OGS-compatible mesh
msh_path_VPF = mesh_path_VPF.joinpath(f"{meshname}.msh")…
(click to toggle)
msh_path_VPF = mesh_path_VPF.joinpath(f"{meshname}.msh")
meshes_volume_VPF = ot.meshes_from_gmsh(
filename=msh_path_VPF, dim=[0, 1], reindex=True, log=False
)
for name, mesh in meshes_volume_VPF.items():
print(f"{name}: {mesh.n_cells} cells")
pv.save_meshio(mesh_path_VPF / f"{name}.vtu", mesh)domain: 104 cells
physical_group_p_right: 1 cells
physical_group_p_top: 1 cells
physical_group_p_left: 1 cells
physical_group_p_bottom: 1 cells
physical_group_PEE1: 4 cells
physical_group_PEE2: 4 cells
physical_group_PEE3: 4 cells
physical_group_PEE4: 4 cells
physical_group_PEE5: 4 cells
physical_group_PEE6: 4 cells
physical_group_PEE7: 4 cells
physical_group_PEE8: 4 cells
physical_group_PEE1a: 4 cells
physical_group_PEE2a: 4 cells
physical_group_PEE3a: 4 cells
physical_group_PEE4a: 4 cells
physical_group_PEE5a: 4 cells
physical_group_PEE6a: 4 cells
physical_group_PEE7a: 4 cells
physical_group_PEE8a: 4 cells
physical_group_DSS1: 2 cells
physical_group_DSS2: 2 cells
physical_group_DSS3: 2 cells
physical_group_DSS4: 2 cells
physical_group_DSS5: 2 cells
physical_group_DSS6: 2 cells
physical_group_DSS7: 2 cells
physical_group_DSS8: 2 cells
physical_group_DSS1a: 2 cells
physical_group_DSS2a: 2 cells
physical_group_DSS3a: 2 cells
physical_group_DSS4a: 2 cells
physical_group_DSS5a: 2 cells
physical_group_DSS6a: 2 cells
physical_group_DSS7a: 2 cells
physical_group_DSS8a: 2 cells
physical_group_Inlet: 1 cells
physical_group_Outlet_R_embeddedFracture: 1 cells
physical_group_Outlet_R_fullFracture: 1 cells
physical_group_Outlet_L_fullFracture: 1 cells
Visualization of boundary meshes
plotter = pv.Plotter()…
(click to toggle)
plotter = pv.Plotter()
for name, mesh in meshes_volume_VPF.items():
if mesh.active_scalars is not None:
plotter.add_mesh(
mesh,
scalars=mesh.active_scalars_name,
cmap="tab20",
show_edges=False,
opacity=0.7,
)
else:
plotter.add_mesh(mesh, show_edges=False, opacity=0.7, label=name)
clean_name = name.replace("physical_group_", "")
center = mesh.center
direction = np.array(center) - np.array([0, 0, 0])
direction[:2] = direction[:2] / (np.linalg.norm(direction[:2]) + 1e-8)
offset = center + 0.025 * direction
plotter.add_point_labels(
[offset], [clean_name], font_size=12, point_size=0, text_color="black"
)
plotter.view_xy()
plotter.enable_parallel_projection()
plotter.show()

Computational domain mesh
Gmsh
msh_file_VPF = mesh_GreatCell_VPF(…
(click to toggle)
msh_file_VPF = mesh_GreatCell_VPF(
lc2=h,
lc=20 * h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path_VPF,
meshname=meshname,
mode="domain",
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 10%] Meshing curve 110 (Line)
Info : [ 10%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 20%] Meshing curve 118 (Line)
Info : [ 20%] Meshing curve 119 (Line)
Info : [ 20%] Meshing curve 120 (Line)
Info : [ 20%] Meshing curve 121 (Line)
Info : [ 20%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 30%] Meshing curve 126 (Line)
Info : [ 30%] Meshing curve 127 (Line)
Info : [ 30%] Meshing curve 128 (Line)
Info : [ 30%] Meshing curve 129 (Line)
Info : [ 30%] Meshing curve 130 (Line)
Info : [ 30%] Meshing curve 131 (Line)
Info : [ 30%] Meshing curve 132 (Line)
Info : [ 30%] Meshing curve 133 (Line)
Info : [ 40%] Meshing curve 134 (Line)
Info : [ 40%] Meshing curve 135 (Line)
Info : [ 40%] Meshing curve 136 (Line)
Info : [ 40%] Meshing curve 137 (Line)
Info : [ 40%] Meshing curve 138 (Line)
Info : [ 40%] Meshing curve 139 (Line)
Info : [ 40%] Meshing curve 140 (Line)
Info : [ 40%] Meshing curve 141 (Line)
Info : [ 40%] Meshing curve 142 (Line)
Info : [ 40%] Meshing curve 143 (Line)
Info : [ 40%] Meshing curve 144 (Line)
Info : [ 50%] Meshing curve 145 (Line)
Info : [ 50%] Meshing curve 146 (Line)
Info : [ 50%] Meshing curve 147 (Line)
Info : [ 50%] Meshing curve 148 (Line)
Info : [ 50%] Meshing curve 149 (Line)
Info : [ 50%] Meshing curve 150 (Line)
Info : [ 50%] Meshing curve 151 (Line)
Info : [ 50%] Meshing curve 152 (Line)
Info : [ 50%] Meshing curve 153 (Line)
Info : [ 50%] Meshing curve 154 (Line)
Info : [ 60%] Meshing curve 155 (Line)
Info : [ 60%] Meshing curve 156 (Line)
Info : [ 60%] Meshing curve 157 (Line)
Info : [ 60%] Meshing curve 158 (Line)
Info : [ 60%] Meshing curve 159 (Line)
Info : [ 60%] Meshing curve 160 (Line)
Info : [ 60%] Meshing curve 161 (Line)
Info : [ 60%] Meshing curve 162 (Line)
Info : [ 60%] Meshing curve 163 (Line)
Info : [ 60%] Meshing curve 164 (Line)
Info : [ 60%] Meshing curve 401 (Circle)
Info : [ 70%] Meshing curve 402 (Circle)
Info : [ 70%] Meshing curve 403 (Circle)
Info : [ 70%] Meshing curve 404 (Circle)
Info : [ 70%] Meshing curve 411 (Circle)
Info : [ 70%] Meshing curve 412 (Circle)
Info : [ 70%] Meshing curve 413 (Circle)
Info : [ 70%] Meshing curve 414 (Circle)
Info : [ 70%] Meshing curve 421 (Circle)
Info : [ 70%] Meshing curve 422 (Circle)
Info : [ 70%] Meshing curve 423 (Circle)
Info : [ 70%] Meshing curve 424 (Circle)
Info : [ 80%] Meshing curve 431 (Circle)
Info : [ 80%] Meshing curve 432 (Circle)
Info : [ 80%] Meshing curve 433 (Circle)
Info : [ 80%] Meshing curve 434 (Circle)
Info : [ 80%] Meshing curve 501 (Line)
Info : [ 80%] Meshing curve 502 (Line)
Info : [ 80%] Meshing curve 503 (Line)
Info : [ 80%] Meshing curve 504 (Line)
Info : [ 80%] Meshing curve 505 (Line)
Info : [ 80%] Meshing curve 506 (Line)
Info : [ 80%] Meshing curve 507 (Line)
Info : [ 90%] Meshing curve 508 (Line)
Info : [ 90%] Meshing curve 509 (Line)
Info : [ 90%] Meshing curve 510 (Line)
Info : [ 90%] Meshing curve 511 (Line)
Info : [ 90%] Meshing curve 512 (Line)
Info : [ 90%] Meshing curve 513 (Line)
Info : [ 90%] Meshing curve 514 (Line)
Info : [ 90%] Meshing curve 515 (Line)
Info : [ 90%] Meshing curve 516 (Line)
Info : [ 90%] Meshing curve 517 (Line)
Info : [ 90%] Meshing curve 518 (Line)
Info : [100%] Meshing curve 519 (Line)
Info : [100%] Meshing curve 520 (Line)
Info : [100%] Meshing curve 521 (Line)
Info : [100%] Meshing curve 522 (Line)
Info : [100%] Meshing curve 523 (Line)
Info : [100%] Meshing curve 524 (Line)
Info : [100%] Meshing curve 525 (Line)
Info : [100%] Meshing curve 526 (Line)
Info : [100%] Meshing curve 5040 (Line)
Info : [100%] Meshing curve 5120 (Line)
Info : Done meshing 1D (Wall 0.028733s, CPU 0.031074s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 701 (Plane, Frontal-Delaunay for Quads)
Info : [ 0%] Blossom: 51 internal 52 closed
Info : [ 0%] Blossom recombination completed (Wall 0.000114533s, CPU 0s): 26 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 10%] Meshing surface 702 (Plane, Frontal-Delaunay for Quads)
Info : [ 10%] Blossom: 79 internal 80 closed
Info : [ 10%] Blossom recombination completed (Wall 0.000161755s, CPU 0.000177s): 40 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 20%] Meshing surface 703 (Plane, Frontal-Delaunay for Quads)
Info : [ 20%] Blossom: 129 internal 130 closed
Info : [ 20%] Blossom recombination completed (Wall 0.000261245s, CPU 0.000282s): 65 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 20%] Meshing surface 704 (Plane, Frontal-Delaunay for Quads)
Info : [ 20%] Blossom recombination completed (Wall 0.00729436s, CPU 0.007592s): 639 quads, 133 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.843335, min Q = 0.363603
Info : [ 30%] Meshing surface 705 (Plane, Frontal-Delaunay for Quads)
Info : [ 30%] Blossom recombination completed (Wall 0.00569474s, CPU 0.005758s): 665 quads, 125 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.844908, min Q = 0.460748
Info : [ 30%] Meshing surface 706 (Plane, Frontal-Delaunay for Quads)
Info : [ 30%] Blossom: 49 internal 50 closed
Info : [ 30%] Blossom recombination completed (Wall 0.000141554s, CPU 0.00016s): 25 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 40%] Meshing surface 707 (Plane, Frontal-Delaunay for Quads)
Info : [ 40%] Blossom: 49 internal 50 closed
Info : [ 40%] Blossom recombination completed (Wall 0.000135836s, CPU 0.000156s): 25 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 40%] Meshing surface 708 (Plane, Frontal-Delaunay for Quads)
Info : [ 40%] Blossom: 1190 internal 132 closed
Info : [ 40%] Blossom recombination completed (Wall 0.00780059s, CPU 0.007792s): 416 quads, 0 triangles, 0 invalid quads, 1 quads with Q < 0.1, avg Q = 0.744994, min Q = 0.0703248
Info : [ 50%] Meshing surface 709 (Plane, Frontal-Delaunay for Quads)
Info : [ 50%] Blossom: 1130 internal 132 closed
Info : [ 50%] Blossom recombination completed (Wall 0.00748149s, CPU 0.00748s): 395 quads, 0 triangles, 0 invalid quads, 3 quads with Q < 0.1, avg Q = 0.762586, min Q = 0.0293523
Info : [ 60%] Meshing surface 710 (Plane, Frontal-Delaunay for Quads)
Info : [ 60%] Blossom: 7 internal 8 closed
Info : [ 60%] Blossom recombination completed (Wall 9.09274e-05s, CPU 0.000109s): 4 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 60%] Meshing surface 711 (Plane, Frontal-Delaunay for Quads)
Info : [ 60%] Blossom: 7 internal 8 closed
Info : [ 60%] Blossom recombination completed (Wall 8.16933e-05s, CPU 0.000101s): 4 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 70%] Meshing surface 712 (Plane, Frontal-Delaunay for Quads)
Info : [ 70%] Blossom: 197 internal 103 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00104155s, CPU 0.001057s): 80 quads, 6 triangles, 0 invalid quads, 6 quads with Q < 0.1, avg Q = 0.564073, min Q = 0.0244822
Info : [ 70%] Meshing surface 713 (Plane, Frontal-Delaunay for Quads)
Info : [ 70%] Blossom: 191 internal 102 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00102087s, CPU 0.001037s): 75 quads, 12 triangles, 0 invalid quads, 8 quads with Q < 0.1, avg Q = 0.52015, min Q = 0.0178184
Info : [ 80%] Meshing surface 714 (Plane, Frontal-Delaunay for Quads)
Info : [ 80%] Blossom: 5 internal 6 closed
Info : [ 80%] Blossom recombination completed (Wall 8.88747e-05s, CPU 0.000109s): 3 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 80%] Meshing surface 715 (Plane, Frontal-Delaunay for Quads)
Info : [ 80%] Blossom: 5 internal 6 closed
Info : [ 80%] Blossom recombination completed (Wall 8.34172e-05s, CPU 0.000101s): 3 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 1, min Q = 1
Info : [ 90%] Meshing surface 716 (Plane, Frontal-Delaunay for Quads)
Info : [ 90%] Blossom: 67 internal 68 closed
Info : [ 90%] Blossom recombination completed (Wall 0.000241124s, CPU 0.000262s): 18 quads, 32 triangles, 0 invalid quads, 4 quads with Q < 0.1, avg Q = 0.275269, min Q = 0.0598336
Info : [ 90%] Meshing surface 717 (Plane, Frontal-Delaunay for Quads)
Info : [ 90%] Blossom: 67 internal 68 closed
Info : [ 90%] Blossom recombination completed (Wall 0.000237679s, CPU 0.000256s): 18 quads, 32 triangles, 0 invalid quads, 4 quads with Q < 0.1, avg Q = 0.275269, min Q = 0.0598336
Info : [100%] Meshing surface 718 (Plane, Frontal-Delaunay for Quads)
Info : [100%] Blossom: 130 internal 130 closed
Info : [100%] Blossom recombination completed (Wall 0.00040325s, CPU 0.000423s): 50 quads, 30 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.144988, min Q = 0.125
Info : Done meshing 2D (Wall 0.0775401s, CPU 0.077577s)
Info : 2786 nodes 3806 elements
Info : Writing '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/GreatCell.msh'
Warning : Cannot apply Blossom: odd number of triangles (1413) in surface 704
Warning : Cannot apply Blossom: odd number of triangles (1455) in surface 705
Warning : ------------------------------
Warning : Mesh generation error summary
Warning : 2 warnings
Warning : 0 errors
Warning : Check the full log for details
Warning : ------------------------------
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/domain.vtu (2921 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Central_sample.vtu (2841 cells)
Saved: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Rubber_sheath.vtu (80 cells)

Convert .msh to an OGS-compatible mesh
msh_path_VPF = mesh_path_VPF.joinpath(f"{meshname}.msh")…
(click to toggle)
msh_path_VPF = mesh_path_VPF.joinpath(f"{meshname}.msh")
meshes_volume_VPF = ot.meshes_from_gmsh(
filename=msh_path_VPF, dim=[2], reindex=True, log=False
)
for name, mesh in meshes_volume_VPF.items():
print(f"{name}: {mesh.n_cells} cells")
pv.save_meshio(mesh_path_VPF.joinpath(f"{name}.vtu"), mesh)domain: 2921 cells
physical_group_Central_sample: 2841 cells
physical_group_Rubber_sheath: 80 cells
Visualization of computational domain mesh
plotter = pv.Plotter()…
(click to toggle)
plotter = pv.Plotter()
for name, mesh in meshes_volume_VPF.items():
if mesh.active_scalars is not None:
plotter.add_mesh(
mesh,
scalars=mesh.active_scalars_name,
cmap="Set1",
show_edges=False,
opacity=0.7,
)
else:
plotter.add_mesh(mesh, show_edges=False, opacity=0.7, label=name)
clean_name = name.replace("physical_group_", "")
center = mesh.center
direction = np.array(center) - np.array([0, 0, 0])
direction[:2] = direction[:2] / (np.linalg.norm(direction[:2]) + 1e-8)
offset = center + 0.025 * direction
plotter.add_point_labels(
[offset], [clean_name], font_size=12, point_size=0, text_color="black"
)
plotter.view_xy()
plotter.enable_parallel_projection()
plotter.show()

mesh_dir = Path(mesh_path_VPF).resolve()…
(click to toggle)
mesh_dir = Path(mesh_path_VPF).resolve()
run(
[
"NodeReordering",
"-i",
str(mesh_dir.joinpath("domain.vtu")),
"-o",
str(mesh_dir.joinpath("domain.vtu")),
],
cwd=mesh_dir,
check=True,
)
physical_groups = [
"physical_group_DSS1.vtu",
"physical_group_DSS1a.vtu",
"physical_group_DSS2.vtu",
"physical_group_DSS2a.vtu",
"physical_group_DSS3.vtu",
"physical_group_DSS3a.vtu",
"physical_group_DSS4.vtu",
"physical_group_DSS4a.vtu",
"physical_group_DSS5.vtu",
"physical_group_DSS5a.vtu",
"physical_group_DSS6.vtu",
"physical_group_DSS6a.vtu",
"physical_group_DSS7.vtu",
"physical_group_DSS7a.vtu",
"physical_group_DSS8.vtu",
"physical_group_DSS8a.vtu",
"physical_group_PEE1.vtu",
"physical_group_PEE1a.vtu",
"physical_group_PEE2.vtu",
"physical_group_PEE2a.vtu",
"physical_group_PEE3.vtu",
"physical_group_PEE3a.vtu",
"physical_group_PEE4.vtu",
"physical_group_PEE4a.vtu",
"physical_group_PEE5.vtu",
"physical_group_PEE5a.vtu",
"physical_group_PEE6.vtu",
"physical_group_PEE6a.vtu",
"physical_group_PEE7.vtu",
"physical_group_PEE7a.vtu",
"physical_group_PEE8.vtu",
"physical_group_PEE8a.vtu",
"physical_group_p_bottom.vtu",
"physical_group_p_left.vtu",
"physical_group_p_right.vtu",
"physical_group_p_top.vtu",
"physical_group_Inlet.vtu",
"physical_group_Outlet_R_embeddedFracture.vtu",
"physical_group_Outlet_R_fullFracture.vtu",
"physical_group_Outlet_L_fullFracture.vtu",
]
group_paths = [str(mesh_dir.joinpath(name)) for name in physical_groups]
run(
[
"identifySubdomains",
"-f",
"-m",
str(mesh_dir.joinpath("domain.vtu")),
"--",
*group_paths,
],
cwd=mesh_dir,
check=True,
)[0] info: Reordering nodes...
[0] info: Method: Reversing order of nodes unless it is considered correct by the OGS6 standard, i.e. such that det(J) > 0, where J is the Jacobian of the global-to-local coordinate transformation.
[0] info: Corrected 1469 elements.
[0] info: VTU file written.
[0] info: Mesh reading time: 0.0183975 s
[0] info: MeshNodeSearcher construction time: 0.000162936 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.405e-06 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000217178 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.71e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1a' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000137588 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.71e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000137188 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.91e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2a' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000135084 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.41e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000134163 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.81e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3a' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000136837 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.21e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000135415 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.31e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4a' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000141945 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.81e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000135324 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.9e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5a' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000136296 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.91e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000137047 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.01e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6a' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000134123 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.41e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000135435 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.31e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7a' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000133642 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.6e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000140433 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.41e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8a' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000134594 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.52e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000134003 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.21e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000131469 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.62e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000136446 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.11e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000137117 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.21e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000138019 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.51e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000142666 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.51e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000134984 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.61e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000136817 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.21e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000138499 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.91e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000138269 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.92e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000135195 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.91e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000135455 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.51e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000140102 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.002e-06 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000134894 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.01e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000136236 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.52e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8a' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000134794 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.81e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_bottom' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000133953 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.31e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_left' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000137178 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.11e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_right' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000133973 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.21e-07 s
[0] info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_top' and it is equal to the newly computed values.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000135364 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.71e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_Inlet' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000136597 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.11e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_Outlet_R_embeddedFracture' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000135875 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.11e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_Outlet_R_fullFracture' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000135044 s
[0] info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.61e-07 s
[0] warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_Outlet_L_fullFracture' and it is not equal to the newly computed values.
[0] info: Overwriting 'bulk_node_ids' property.
[0] info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000137257 s
[0] info: identifySubdomains time: 0.00588047 s
[0] info: writing time: 0.0275762 s
[0] info: Entire run time: -2.26792e+06 s
CompletedProcess(args=['identifySubdomains', '-f', '-m', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/domain.vtu', '--', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS1.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS1a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS2.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS2a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS3.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS3a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS4.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS4a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS5.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS5a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS6.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS6a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS7.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS7a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS8.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS8a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE1.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE1a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE2.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE2a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE3.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE3a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE4.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE4a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE5.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE5a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE6.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE6a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE7.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE7a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE8.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE8a.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_bottom.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_left.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_right.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_top.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Inlet.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_R_embeddedFracture.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_R_fullFracture.vtu', '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_L_fullFracture.vtu'], returncode=0)
Half fractured sample ($\texttt{M}_{2b}$)
In this section, all input data and boundary conditions provided in $\texttt{M}_{2b}$, except a single wing fracture, defined as $\Gamma = \left[0, 0.04\right] \times \{0\}$, is considered.
Run the simulation
Inputs
# Times for load curves…
(click to toggle)
# Times for load curves
times = "0.0 1000. 3500"
simulation_end_time = 3500.0
n_fracture_p_ncs = 3
model_type = "M2b"
output_prefix = "M2b_VPF"
# Load
PEE_load_values = {
"A": [10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6, 9.95e6],
"B": [7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6],
"C": [1.0e6, 3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6],
}
# Project file
prj_file = Path("M2_VPF.prj")
prj = ot.Project(input_file=prj_file, output_file=Path(out_dir, f"{output_prefix}.prj"))Run OGS
sing_ogs_model = SingleOGSModel(
model=prj,
out_dir=out_dir,
mesh_path=mesh_path_VPF,
output_prefix=output_prefix,
method="VPF",
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
materials=materials,
n_mpi=2,
)
vtu_files_dict_embedded = sing_ogs_model.run_simulations_with_fracture(
times=times,
base_project_file=prj_file,
mesh_path=mesh_path_VPF,
load_cases=PEE_load_values,
material_names=material_names,
materials=materials,
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
output_prefix=output_prefix,
out_dir=out_dir,
method="VPF",
crack_type="half",
fracture_model_type="VolumetricDeviatoric",
mesh_size=h,
n_mpi=2,
)mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2b_VPF.prj
Modifying mesh for VPF method at /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF with crack type: half and h1: 0.001
[0] info: Mesh 'domain' read: 2785 nodes, 2921 elements.
[0] info: Property global_node_ids is added to mesh domain
[0] info: Property global_element_ids is added to mesh domain
[0] info: Write the mesh into METIS input file.
[0] info: Total runtime: 0.0128909 s.
[0] info: Total CPU time: 0.012778 s.
[0] info: Mesh 'domain' read: 2785 nodes, 2921 elements.
[0] info: Property global_node_ids is added to mesh domain
[0] info: Property global_element_ids is added to mesh domain
[0] info: METIS is running ...
[0] info: Path to mpmetis is:
[0] info: Running: mpmetis -gtype=nodal "/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/domain.mesh" 2
******************************************************************************
METIS 5.2.1 Copyright 1998-22, Regents of the University of Minnesota
(HEAD: unknown, Built on: Sep 26 2025, 11:30:11)
size of idx_t: 64bits, real_t: 32bits, idx_t *: 64bits
Mesh Information ------------------------------------------------------------
Name: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/domain.mesh, #Elements: 2921, #Nodes: 2785, #Parts: 2
Options ---------------------------------------------------------------------
ptype=kway, objtype=cut, ctype=shem, rtype=greedy, iptype=metisrb
dbglvl=0, ufactor=1.030, minconn=NO, contig=NO, nooutput=NO
seed=-1, niter=10, ncuts=1
gtype=nodal, ncommon=1, niter=10, ncuts=1
Direct k-way Partitioning ---------------------------------------------------
- Edgecut: 119.
Timing Information ----------------------------------------------------------
I/O: 0.001 sec
Partitioning: 0.002 sec (METIS time)
Reporting: -0.000 sec
Memory Information ----------------------------------------------------------
Max memory used: 0.902 MB
rusage.ru_maxrss: 108.922 MB
proc/self/stat/VmPeak: 25.488 MB
******************************************************************************
[0] info: Partitioning the mesh in the node wise way ...
[0] info: partitionMesh(): Partition IDs per element computed in 0.000122766 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.34e-05 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 0.000177819 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 0.000100212 s
[0] info: partitionMesh(): distribute elements into partitions took 8.3627e-05 s
[0] info: partitionMesh(): determine / append ghost nodes took 1.9259e-05 s
[0] info: partitionMesh(): markDuplicateGhostCells took 7.02e-07 s
[0] info: Partitioning other meshes according to the main mesh partitions.
[0] info: Mesh 'physical_group_DSS1' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS1.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS1
[0] info: Property global_element_ids is added to mesh physical_group_DSS1
[0] info: partitionMesh(): Partition IDs per element computed in 2.6e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.8e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.6e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.7e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.71e-07 s
[0] info: Mesh 'physical_group_DSS1a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS1a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS1a
[0] info: Property global_element_ids is added to mesh physical_group_DSS1a
[0] info: partitionMesh(): Partition IDs per element computed in 2.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.1e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.51e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.31e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.5e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.9e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.81e-07 s
[0] info: Mesh 'physical_group_DSS2' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS2.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS2
[0] info: Property global_element_ids is added to mesh physical_group_DSS2
[0] info: partitionMesh(): Partition IDs per element computed in 2.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.11e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 9e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 3.5e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.71e-07 s
[0] info: Mesh 'physical_group_DSS2a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS2a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS2a
[0] info: Property global_element_ids is added to mesh physical_group_DSS2a
[0] info: partitionMesh(): Partition IDs per element computed in 2.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.4e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.2e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_DSS3' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS3.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS3
[0] info: Property global_element_ids is added to mesh physical_group_DSS3
[0] info: partitionMesh(): Partition IDs per element computed in 2.3e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.8e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.5e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.3e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.31e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_DSS3a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS3a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS3a
[0] info: Property global_element_ids is added to mesh physical_group_DSS3a
[0] info: partitionMesh(): Partition IDs per element computed in 2.31e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.8e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.11e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.31e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_DSS4' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS4.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS4
[0] info: Property global_element_ids is added to mesh physical_group_DSS4
[0] info: partitionMesh(): Partition IDs per element computed in 2.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.6e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 9.1e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 4.7e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.11e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_DSS4a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS4a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS4a
[0] info: Property global_element_ids is added to mesh physical_group_DSS4a
[0] info: partitionMesh(): Partition IDs per element computed in 2.3e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.1e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.3e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 9.1e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 3.3e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2e-07 s
[0] info: Mesh 'physical_group_DSS5' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS5.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS5
[0] info: Property global_element_ids is added to mesh physical_group_DSS5
[0] info: partitionMesh(): Partition IDs per element computed in 2.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.1e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.71e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.9e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_DSS5a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS5a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS5a
[0] info: Property global_element_ids is added to mesh physical_group_DSS5a
[0] info: partitionMesh(): Partition IDs per element computed in 3.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.7e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.31e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 9e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 3.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 4.41e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_DSS6' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS6.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS6
[0] info: Property global_element_ids is added to mesh physical_group_DSS6
[0] info: partitionMesh(): Partition IDs per element computed in 2.3e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.7e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.11e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.3e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.81e-07 s
[0] info: Mesh 'physical_group_DSS6a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS6a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS6a
[0] info: Property global_element_ids is added to mesh physical_group_DSS6a
[0] info: partitionMesh(): Partition IDs per element computed in 3.11e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.81e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.2e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.8e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.71e-07 s
[0] info: Mesh 'physical_group_DSS7' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS7.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS7
[0] info: Property global_element_ids is added to mesh physical_group_DSS7
[0] info: partitionMesh(): Partition IDs per element computed in 2.61e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.8e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.01e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 9e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 3.3e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.81e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_DSS7a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS7a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS7a
[0] info: Property global_element_ids is added to mesh physical_group_DSS7a
[0] info: partitionMesh(): Partition IDs per element computed in 2.51e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 9.1e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 3.7e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.7e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_DSS8' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS8.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS8
[0] info: Property global_element_ids is added to mesh physical_group_DSS8
[0] info: partitionMesh(): Partition IDs per element computed in 2.3e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.9e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 8e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 3.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.7e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2e-07 s
[0] info: Mesh 'physical_group_DSS8a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS8a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS8a
[0] info: Property global_element_ids is added to mesh physical_group_DSS8a
[0] info: partitionMesh(): Partition IDs per element computed in 2.5e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.1e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.1e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 9e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 3.5e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.6e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_PEE1' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE1.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE1
[0] info: Property global_element_ids is added to mesh physical_group_PEE1
[0] info: partitionMesh(): Partition IDs per element computed in 2.9e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 4.11e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.61e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.61e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.6e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2e-07 s
[0] info: Mesh 'physical_group_PEE1a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE1a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE1a
[0] info: Property global_element_ids is added to mesh physical_group_PEE1a
[0] info: partitionMesh(): Partition IDs per element computed in 3e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.51e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.31e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.4e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.5e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.91e-07 s
[0] info: Mesh 'physical_group_PEE2' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE2.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE2
[0] info: Property global_element_ids is added to mesh physical_group_PEE2
[0] info: partitionMesh(): Partition IDs per element computed in 2.7e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.81e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.9e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.71e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.5e-07 s
[0] info: Mesh 'physical_group_PEE2a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE2a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE2a
[0] info: Property global_element_ids is added to mesh physical_group_PEE2a
[0] info: partitionMesh(): Partition IDs per element computed in 2.7e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.1e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.81e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.51e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.1e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.8e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_PEE3' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE3.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE3
[0] info: Property global_element_ids is added to mesh physical_group_PEE3
[0] info: partitionMesh(): Partition IDs per element computed in 2.9e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.11e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.51e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.7e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.11e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.9e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_PEE3a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE3a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE3a
[0] info: Property global_element_ids is added to mesh physical_group_PEE3a
[0] info: partitionMesh(): Partition IDs per element computed in 2.71e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.41e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.41e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.1e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_PEE4' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE4.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE4
[0] info: Property global_element_ids is added to mesh physical_group_PEE4
[0] info: partitionMesh(): Partition IDs per element computed in 2.7e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.21e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.81e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.8e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_PEE4a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE4a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE4a
[0] info: Property global_element_ids is added to mesh physical_group_PEE4a
[0] info: partitionMesh(): Partition IDs per element computed in 2.61e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.71e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.3e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.6e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 7.92e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3e-07 s
[0] info: Mesh 'physical_group_PEE5' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE5.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE5
[0] info: Property global_element_ids is added to mesh physical_group_PEE5
[0] info: partitionMesh(): Partition IDs per element computed in 3.11e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.51e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.5e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.4e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.1e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 1.0416e-05 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2.91e-07 s
[0] info: Mesh 'physical_group_PEE5a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE5a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE5a
[0] info: Property global_element_ids is added to mesh physical_group_PEE5a
[0] info: partitionMesh(): Partition IDs per element computed in 2.6e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.5e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.51e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.81e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.8e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_PEE6' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE6.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE6
[0] info: Property global_element_ids is added to mesh physical_group_PEE6
[0] info: partitionMesh(): Partition IDs per element computed in 2.71e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.31e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.2e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_PEE6a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE6a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE6a
[0] info: Property global_element_ids is added to mesh physical_group_PEE6a
[0] info: partitionMesh(): Partition IDs per element computed in 2.6e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.3e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.41e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.4e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.71e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_PEE7' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE7.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE7
[0] info: Property global_element_ids is added to mesh physical_group_PEE7
[0] info: partitionMesh(): Partition IDs per element computed in 2.71e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.11e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.81e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.1e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_PEE7a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE7a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE7a
[0] info: Property global_element_ids is added to mesh physical_group_PEE7a
[0] info: partitionMesh(): Partition IDs per element computed in 2.8e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.71e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.01e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.41e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.91e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2e-07 s
[0] info: Mesh 'physical_group_PEE8' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE8.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE8
[0] info: Property global_element_ids is added to mesh physical_group_PEE8
[0] info: partitionMesh(): Partition IDs per element computed in 2.6e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.31e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.31e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.8e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.81e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.91e-07 s
[0] info: Mesh 'physical_group_PEE8a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE8a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE8a
[0] info: Property global_element_ids is added to mesh physical_group_PEE8a
[0] info: partitionMesh(): Partition IDs per element computed in 2.71e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.11e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.61e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.3e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.71e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2.4e-07 s
[0] info: Mesh 'physical_group_p_bottom' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_bottom.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_bottom
[0] info: Property global_element_ids is added to mesh physical_group_p_bottom
[0] info: partitionMesh(): Partition IDs per element computed in 2.1e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 8e-08 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.2e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 7e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 1.71e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.21e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.91e-07 s
[0] info: Mesh 'physical_group_p_left' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_left.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_left
[0] info: Property global_element_ids is added to mesh physical_group_p_left
[0] info: partitionMesh(): Partition IDs per element computed in 2e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.3e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 6e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2.5e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.9e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_p_right' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_right.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_right
[0] info: Property global_element_ids is added to mesh physical_group_p_right
[0] info: partitionMesh(): Partition IDs per element computed in 1.81e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 8e-08 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.41e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 6e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2.21e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.7e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_p_top' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_top.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_top
[0] info: Property global_element_ids is added to mesh physical_group_p_top
[0] info: partitionMesh(): Partition IDs per element computed in 2e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 7e-08 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.1e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 7.1e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 1.9e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.6e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_Inlet' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Inlet.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Inlet
[0] info: Property global_element_ids is added to mesh physical_group_Inlet
[0] info: partitionMesh(): Partition IDs per element computed in 2.11e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.7e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.81e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 7e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.8e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_Outlet_R_embeddedFracture' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_R_embeddedFracture.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Outlet_R_embeddedFracture
[0] info: Property global_element_ids is added to mesh physical_group_Outlet_R_embeddedFracture
[0] info: partitionMesh(): Partition IDs per element computed in 2.21e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.4e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.51e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 8e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2.1e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.9e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_Outlet_R_fullFracture' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_R_fullFracture.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Outlet_R_fullFracture
[0] info: Property global_element_ids is added to mesh physical_group_Outlet_R_fullFracture
[0] info: partitionMesh(): Partition IDs per element computed in 2.9e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.4e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 7e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2.01e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.7e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_Outlet_L_fullFracture' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_L_fullFracture.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Outlet_L_fullFracture
[0] info: Property global_element_ids is added to mesh physical_group_Outlet_L_fullFracture
[0] info: partitionMesh(): Partition IDs per element computed in 2.01e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.1e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.81e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 8e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2.01e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Writing the partitions data into binary files took 0.000956687 s
[0] info: Total runtime: 0.0984708 s.
[0] info: Total CPU time: 0.04857 s.
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2b_VPF_A.prj
============================================================
Running simulation for load case: A with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2b_VPF_A.prj
Status: finished successfully.
Execution took 9.751033306121826 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2b_VPF_A.prj
Status: finished successfully.
Execution took 9.710178852081299 s
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2b_VPF_B.prj
============================================================
Running simulation for load case: B with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2b_VPF_B.prj
Status: finished successfully.
Execution took 9.444863080978394 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2b_VPF_B.prj
Status: finished successfully.
Execution took 9.811509609222412 s
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2b_VPF_C.prj
============================================================
Running simulation for load case: C with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2b_VPF_C.prj
Status: finished successfully.
Execution took 8.70984172821045 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2b_VPF_C.prj
Status: finished successfully.
Execution took 8.848543405532837 s
Post-processing
plotter = Plotter(…
(click to toggle)
plotter = Plotter(
output_dir=out_dir,
save_extracted_data=True,
)
data_dir = Path("external_data")
external_data = Plotter.load_external_data(data_dir, benchmark_tag="M2b")
plotter.plot_volumetric_strain_vs_angle(
vtu_files_dict_embedded,
model_type="M2b",
ylim_range=[-7.5, 2.5],
layout="single",
external_data=external_data["strain"],
)
plotter.plot_field_variables(vtu_files_dict_embedded)[0] info: Reading meshes took 0.012171774999999885 s
[0] info: Collection of 2921 regular elements and computing element map took 2.1431999999821372e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.0540000000023326e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.001206033999999967 s
[0] info: Reset nodes in regular elements took 0.0057122170000001304 s
[0] info: creation of merged mesh took 0.0005069570000000301 s
[0] info: merge properties into merged mesh took 0.0003632590000002267 s
[0] info: writing mesh took 0.012914797999999728 s
[0] info: Reading meshes took 0.012431287000000069 s
[0] info: Collection of 2921 regular elements and computing element map took 2.8573999999892408e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 4.526999999976411e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.001327416999999942 s
[0] info: Reset nodes in regular elements took 0.005604764999999956 s
[0] info: creation of merged mesh took 0.0005358000000001972 s
[0] info: merge properties into merged mesh took 0.0003622880000000439 s
[0] info: writing mesh took 0.012929640000000076 s
[0] info: Reading meshes took 0.010343079999999949 s
[0] info: Collection of 2921 regular elements and computing element map took 2.0831000000054445e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.4550000000965753e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0012053629999999593 s
[0] info: Reset nodes in regular elements took 0.005632145999999949 s
[0] info: creation of merged mesh took 0.0005001860000000136 s
[0] info: merge properties into merged mesh took 0.0003623379999999621 s
[0] info: writing mesh took 0.013222743999999897 s
[0] info: Reading meshes took 0.008917244000000046 s
[0] info: Collection of 2921 regular elements and computing element map took 2.060099999989795e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.1649999998606404e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.001212733999999882 s
[0] info: Reset nodes in regular elements took 0.005601178999999901 s
[0] info: creation of merged mesh took 0.0005587849999999506 s
[0] info: merge properties into merged mesh took 0.0003622070000000033 s
[0] info: writing mesh took 0.012066906000000044 s
[0] info: Reading meshes took 0.008571190000000062 s
[0] info: Collection of 2921 regular elements and computing element map took 2.0110000000128636e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.2240000000793856e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0012098000000000386 s
[0] info: Reset nodes in regular elements took 0.005734601000000117 s
[0] info: creation of merged mesh took 0.0005070469999999272 s
[0] info: merge properties into merged mesh took 0.00036386999999993286 s
[0] info: writing mesh took 0.012771752000000136 s
[0] info: Reading meshes took 0.012101918999999794 s
[0] info: Collection of 2921 regular elements and computing element map took 2.897399999990391e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.385000000077909e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0012064340000002005 s
[0] info: Reset nodes in regular elements took 0.005614028999999743 s
[0] info: creation of merged mesh took 0.0005015379999999681 s
[0] info: merge properties into merged mesh took 0.000363329999999884 s
[0] info: writing mesh took 0.013164104999999982 s

Load: A, File 0, Benchmark: M2b, Material: Gneiss
[0] info: Reading meshes took 0.011337692999999982 s
[0] info: Collection of 2921 regular elements and computing element map took 2.074199999979598e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.405000000178404e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0012047119999998301 s
[0] info: Reset nodes in regular elements took 0.005743865000000126 s
[0] info: creation of merged mesh took 0.0004960899999999491 s
[0] info: merge properties into merged mesh took 0.00037017999999999773 s
[0] info: writing mesh took 0.015317080999999844 s

Load: A, File 1, Benchmark: M2b, Material: Greywacke
[0] info: Reading meshes took 0.01238669899999989 s
[0] info: Collection of 2921 regular elements and computing element map took 2.9564999999953656e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 4.7169999999319145e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0012152379999998963 s
[0] info: Reset nodes in regular elements took 0.005458543999999899 s
[0] info: creation of merged mesh took 0.0005021289999997958 s
[0] info: merge properties into merged mesh took 0.0003629690000002128 s
[0] info: writing mesh took 0.013518852000000026 s

Load: B, File 0, Benchmark: M2b, Material: Gneiss
[0] info: Reading meshes took 0.010637023999999995 s
[0] info: Collection of 2921 regular elements and computing element map took 2.0962000000013248e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.304999999897973e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0012062139999999832 s
[0] info: Reset nodes in regular elements took 0.005692808000000049 s
[0] info: creation of merged mesh took 0.0005000559999999155 s
[0] info: merge properties into merged mesh took 0.000376380000000065 s
[0] info: writing mesh took 0.01481965900000004 s

Load: B, File 1, Benchmark: M2b, Material: Greywacke
[0] info: Reading meshes took 0.01115877300000001 s
[0] info: Collection of 2921 regular elements and computing element map took 2.09819999998917e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.215000000000856e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0012099589999998717 s
[0] info: Reset nodes in regular elements took 0.005624495000000174 s
[0] info: creation of merged mesh took 0.0005091700000001698 s
[0] info: merge properties into merged mesh took 0.0003638299999999539 s
[0] info: writing mesh took 0.015139422000000069 s

Load: C, File 0, Benchmark: M2b, Material: Gneiss
[0] info: Reading meshes took 0.009969804999999887 s
[0] info: Collection of 2921 regular elements and computing element map took 2.7030000000038967e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.324999999998468e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.001212684000000186 s
[0] info: Reset nodes in regular elements took 0.0056488610000000605 s
[0] info: creation of merged mesh took 0.0006077590000002075 s
[0] info: merge properties into merged mesh took 0.0003652819999999668 s
[0] info: writing mesh took 0.01645169600000007 s

Load: C, File 1, Benchmark: M2b, Material: Greywacke
[0] info: Reading meshes took 0.007909850999999968 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9530000000100856e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 2.9939999999228917e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011276749999999947 s
[0] info: Reset nodes in regular elements took 0.005222145999999928 s
[0] info: creation of merged mesh took 0.0004576919999998985 s
[0] info: merge properties into merged mesh took 0.00034487100000002435 s
[0] info: writing mesh took 0.009996606000000074 s

Fully fractured sample ($\texttt{M}_{2a}$)
In this benchmark, 2D plane strain numerical simulations are performed to establish a baseline model for assessing the impact of fracture orientation on strain distribution under poly-axial loading. A planar fracture within the specimen, $\Gamma = \left[-0.094, 0.094\right] \times \{0\}$, is considered, under poly-axial loading applied on PEE’s and DSS’s
Run the simulation
Inputs
# Times for load curves…
(click to toggle)
# Times for load curves
times = "0.0 1000. 3500"
simulation_end_time = 3500.0
n_fracture_p_ncs = 3
model_type = "M2a"
output_prefix = "M2a_VPF"
# Load
PEE_load_values = {
"A": [10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6, 9.95e6],
"B": [7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6],
"C": [1.0e6, 3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6],
}
# Project file
prj_file = Path("M2_VPF.prj")
prj = ot.Project(input_file=prj_file, output_file=Path(out_dir, f"{output_prefix}.prj"))Run OGS
# Create SingleOGSModel…
(click to toggle)
# Create SingleOGSModel
sing_ogs_model = SingleOGSModel(
model=prj,
out_dir=out_dir,
mesh_path=mesh_path_VPF,
output_prefix=output_prefix,
method="VPF",
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
materials=materials,
n_mpi=2,
)
# Run simulations
vtu_files_dict_full = sing_ogs_model.run_simulations_with_fracture(
times=times,
base_project_file=prj_file,
mesh_path=mesh_path_VPF,
load_cases=PEE_load_values,
material_names=material_names,
materials=materials,
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
output_prefix=output_prefix,
out_dir=out_dir,
method="VPF",
crack_type="full",
fracture_model_type="VolumetricDeviatoric",
mesh_size=h,
n_mpi=2,
)mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2a_VPF.prj
Modifying mesh for VPF method at /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF with crack type: full and h1: 0.001
[0] info: Mesh 'domain' read: 2785 nodes, 2921 elements.
[0] info: Property global_node_ids is added to mesh domain
[0] info: Property global_element_ids is added to mesh domain
[0] info: Write the mesh into METIS input file.
[0] info: Total runtime: 0.0119891 s.
[0] info: Total CPU time: 0.011829 s.
[0] info: Mesh 'domain' read: 2785 nodes, 2921 elements.
[0] info: Property global_node_ids is added to mesh domain
[0] info: Property global_element_ids is added to mesh domain
[0] info: METIS is running ...
[0] info: Path to mpmetis is:
[0] info: Running: mpmetis -gtype=nodal "/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/domain.mesh" 2
******************************************************************************
METIS 5.2.1 Copyright 1998-22, Regents of the University of Minnesota
(HEAD: unknown, Built on: Sep 26 2025, 11:30:11)
size of idx_t: 64bits, real_t: 32bits, idx_t *: 64bits
Mesh Information ------------------------------------------------------------
Name: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/domain.mesh, #Elements: 2921, #Nodes: 2785, #Parts: 2
Options ---------------------------------------------------------------------
ptype=kway, objtype=cut, ctype=shem, rtype=greedy, iptype=metisrb
dbglvl=0, ufactor=1.030, minconn=NO, contig=NO, nooutput=NO
seed=-1, niter=10, ncuts=1
gtype=nodal, ncommon=1, niter=10, ncuts=1
Direct k-way Partitioning ---------------------------------------------------
- Edgecut: 119.
Timing Information ----------------------------------------------------------
I/O: 0.001 sec
Partitioning: 0.002 sec (METIS time)
Reporting: 0.000 sec
Memory Information ----------------------------------------------------------
Max memory used: 0.902 MB
rusage.ru_maxrss: 109.027 MB
proc/self/stat/VmPeak: 25.488 MB
******************************************************************************
[0] info: Partitioning the mesh in the node wise way ...
[0] info: partitionMesh(): Partition IDs per element computed in 0.00015849 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.944e-05 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 0.00024427 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 0.000155635 s
[0] info: partitionMesh(): distribute elements into partitions took 9.7628e-05 s
[0] info: partitionMesh(): determine / append ghost nodes took 1.9159e-05 s
[0] info: partitionMesh(): markDuplicateGhostCells took 7.31e-07 s
[0] info: Partitioning other meshes according to the main mesh partitions.
[0] info: Mesh 'physical_group_DSS1' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS1.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS1
[0] info: Property global_element_ids is added to mesh physical_group_DSS1
[0] info: partitionMesh(): Partition IDs per element computed in 2.7e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.51e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.8e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.9e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_DSS1a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS1a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS1a
[0] info: Property global_element_ids is added to mesh physical_group_DSS1a
[0] info: partitionMesh(): Partition IDs per element computed in 2.61e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.41e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.31e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.4e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 4.2e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_DSS2' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS2.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS2
[0] info: Property global_element_ids is added to mesh physical_group_DSS2
[0] info: partitionMesh(): Partition IDs per element computed in 2.71e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.9e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.11e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.6e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.8e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_DSS2a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS2a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS2a
[0] info: Property global_element_ids is added to mesh physical_group_DSS2a
[0] info: partitionMesh(): Partition IDs per element computed in 2.7e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.41e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.61e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 4.01e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_DSS3' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS3.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS3
[0] info: Property global_element_ids is added to mesh physical_group_DSS3
[0] info: partitionMesh(): Partition IDs per element computed in 2.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.71e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.61e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.4e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.7e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.71e-07 s
[0] info: Mesh 'physical_group_DSS3a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS3a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS3a
[0] info: Property global_element_ids is added to mesh physical_group_DSS3a
[0] info: partitionMesh(): Partition IDs per element computed in 2.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.9e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.2e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 9e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 3.6e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.91e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_DSS4' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS4.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS4
[0] info: Property global_element_ids is added to mesh physical_group_DSS4
[0] info: partitionMesh(): Partition IDs per element computed in 2.5e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.9e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.61e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.5e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.71e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.81e-07 s
[0] info: Mesh 'physical_group_DSS4a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS4a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS4a
[0] info: Property global_element_ids is added to mesh physical_group_DSS4a
[0] info: partitionMesh(): Partition IDs per element computed in 2.51e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.9e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.31e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 9e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 3.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.7e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_DSS5' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS5.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS5
[0] info: Property global_element_ids is added to mesh physical_group_DSS5
[0] info: partitionMesh(): Partition IDs per element computed in 2.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.1e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 8e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 3.8e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.6e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_DSS5a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS5a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS5a
[0] info: Property global_element_ids is added to mesh physical_group_DSS5a
[0] info: partitionMesh(): Partition IDs per element computed in 2.61e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.1e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.6e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.41e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_DSS6' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS6.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS6
[0] info: Property global_element_ids is added to mesh physical_group_DSS6
[0] info: partitionMesh(): Partition IDs per element computed in 2.5e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.8e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.01e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.4e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.91e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_DSS6a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS6a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS6a
[0] info: Property global_element_ids is added to mesh physical_group_DSS6a
[0] info: partitionMesh(): Partition IDs per element computed in 2.5e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.6e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.1e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.41e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.4e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.2e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.81e-07 s
[0] info: Mesh 'physical_group_DSS7' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS7.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS7
[0] info: Property global_element_ids is added to mesh physical_group_DSS7
[0] info: partitionMesh(): Partition IDs per element computed in 2.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.8e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_DSS7a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS7a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS7a
[0] info: Property global_element_ids is added to mesh physical_group_DSS7a
[0] info: partitionMesh(): Partition IDs per element computed in 2.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.7e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.61e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.4e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_DSS8' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS8.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS8
[0] info: Property global_element_ids is added to mesh physical_group_DSS8
[0] info: partitionMesh(): Partition IDs per element computed in 2.6e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.71e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.01e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.4e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.6e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.71e-07 s
[0] info: Mesh 'physical_group_DSS8a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS8a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS8a
[0] info: Property global_element_ids is added to mesh physical_group_DSS8a
[0] info: partitionMesh(): Partition IDs per element computed in 2.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.8e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.71e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_PEE1' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE1.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE1
[0] info: Property global_element_ids is added to mesh physical_group_PEE1
[0] info: partitionMesh(): Partition IDs per element computed in 3.21e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.71e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.9e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.5e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.61e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_PEE1a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE1a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE1a
[0] info: Property global_element_ids is added to mesh physical_group_PEE1a
[0] info: partitionMesh(): Partition IDs per element computed in 3.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.6e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.5e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.41e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_PEE2' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE2.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE2
[0] info: Property global_element_ids is added to mesh physical_group_PEE2
[0] info: partitionMesh(): Partition IDs per element computed in 2.8e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.71e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.71e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.41e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_PEE2a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE2a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE2a
[0] info: Property global_element_ids is added to mesh physical_group_PEE2a
[0] info: partitionMesh(): Partition IDs per element computed in 2.8e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.5e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.41e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_PEE3' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE3.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE3
[0] info: Property global_element_ids is added to mesh physical_group_PEE3
[0] info: partitionMesh(): Partition IDs per element computed in 2.8e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.9e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_PEE3a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE3a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE3a
[0] info: Property global_element_ids is added to mesh physical_group_PEE3a
[0] info: partitionMesh(): Partition IDs per element computed in 3.3e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.6e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.11e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.7e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.71e-07 s
[0] info: Mesh 'physical_group_PEE4' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE4.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE4
[0] info: Property global_element_ids is added to mesh physical_group_PEE4
[0] info: partitionMesh(): Partition IDs per element computed in 3.11e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.31e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.8e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_PEE4a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE4a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE4a
[0] info: Property global_element_ids is added to mesh physical_group_PEE4a
[0] info: partitionMesh(): Partition IDs per element computed in 3.31e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.9e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.3e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.41e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 8.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2.81e-07 s
[0] info: Mesh 'physical_group_PEE5' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE5.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE5
[0] info: Property global_element_ids is added to mesh physical_group_PEE5
[0] info: partitionMesh(): Partition IDs per element computed in 3.1e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 4.31e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.4e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.71e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 8.32e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2.7e-07 s
[0] info: Mesh 'physical_group_PEE5a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE5a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE5a
[0] info: Property global_element_ids is added to mesh physical_group_PEE5a
[0] info: partitionMesh(): Partition IDs per element computed in 2.91e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.4e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.31e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.5e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.7e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.81e-07 s
[0] info: Mesh 'physical_group_PEE6' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE6.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE6
[0] info: Property global_element_ids is added to mesh physical_group_PEE6
[0] info: partitionMesh(): Partition IDs per element computed in 2.8e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.51e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.9e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.4e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.81e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 4.01e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_PEE6a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE6a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE6a
[0] info: Property global_element_ids is added to mesh physical_group_PEE6a
[0] info: partitionMesh(): Partition IDs per element computed in 2.81e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.6e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.61e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_PEE7' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE7.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE7
[0] info: Property global_element_ids is added to mesh physical_group_PEE7
[0] info: partitionMesh(): Partition IDs per element computed in 3.01e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.7e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.81e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.21e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.81e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_PEE7a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE7a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE7a
[0] info: Property global_element_ids is added to mesh physical_group_PEE7a
[0] info: partitionMesh(): Partition IDs per element computed in 3.01e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.21e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.5e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.2e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.91e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_PEE8' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE8.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE8
[0] info: Property global_element_ids is added to mesh physical_group_PEE8
[0] info: partitionMesh(): Partition IDs per element computed in 2.7e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.61e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.7e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.81e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.61e-07 s
[0] info: Mesh 'physical_group_PEE8a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE8a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE8a
[0] info: Property global_element_ids is added to mesh physical_group_PEE8a
[0] info: partitionMesh(): Partition IDs per element computed in 2.91e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.3e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.01e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.41e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.71e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_p_bottom' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_bottom.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_bottom
[0] info: Property global_element_ids is added to mesh physical_group_p_bottom
[0] info: partitionMesh(): Partition IDs per element computed in 1.9e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 9e-08 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.31e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 8e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_p_left' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_left.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_left
[0] info: Property global_element_ids is added to mesh physical_group_p_left
[0] info: partitionMesh(): Partition IDs per element computed in 2.21e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 3.81e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 7e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 1.7e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 4.01e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_p_right' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_right.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_right
[0] info: Property global_element_ids is added to mesh physical_group_p_right
[0] info: partitionMesh(): Partition IDs per element computed in 2.21e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.1e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.8e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 7e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.21e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_p_top' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_top.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_top
[0] info: Property global_element_ids is added to mesh physical_group_p_top
[0] info: partitionMesh(): Partition IDs per element computed in 2.91e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 9e-08 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 6e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2.01e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 4e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.81e-07 s
[0] info: Mesh 'physical_group_Inlet' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Inlet.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Inlet
[0] info: Property global_element_ids is added to mesh physical_group_Inlet
[0] info: partitionMesh(): Partition IDs per element computed in 2.11e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.01e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 9e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2.01e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.8e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_Outlet_R_embeddedFracture' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_R_embeddedFracture.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Outlet_R_embeddedFracture
[0] info: Property global_element_ids is added to mesh physical_group_Outlet_R_embeddedFracture
[0] info: partitionMesh(): Partition IDs per element computed in 3.1e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.5e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.1e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 9e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2.3e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.9e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_Outlet_R_fullFracture' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_R_fullFracture.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Outlet_R_fullFracture
[0] info: Property global_element_ids is added to mesh physical_group_Outlet_R_fullFracture
[0] info: partitionMesh(): Partition IDs per element computed in 2e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.41e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 2.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 4.11e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_Outlet_L_fullFracture' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_L_fullFracture.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Outlet_L_fullFracture
[0] info: Property global_element_ids is added to mesh physical_group_Outlet_L_fullFracture
[0] info: partitionMesh(): Partition IDs per element computed in 2e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.9e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.41e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 2e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.8e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Writing the partitions data into binary files took 0.00148708 s
[0] info: Total runtime: 0.100791 s.
[0] info: Total CPU time: 0.050871 s.
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2a_VPF_A.prj
============================================================
Running simulation for load case: A with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2a_VPF_A.prj
Status: finished successfully.
Execution took 11.099642992019653 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2a_VPF_A.prj
Status: finished successfully.
Execution took 10.911139011383057 s
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2a_VPF_B.prj
============================================================
Running simulation for load case: B with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2a_VPF_B.prj
Status: finished successfully.
Execution took 10.178330898284912 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2a_VPF_B.prj
Status: finished successfully.
Execution took 10.421458959579468 s
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2a_VPF_C.prj
============================================================
Running simulation for load case: C with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2a_VPF_C.prj
Status: finished successfully.
Execution took 10.199966192245483 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/M2a_VPF_C.prj
Status: finished successfully.
Execution took 10.467331647872925 s
Post-processing
plotter = Plotter(…
(click to toggle)
plotter = Plotter(
output_dir=out_dir,
save_extracted_data=True,
)
data_dir = Path("external_data")
external_data = Plotter.load_external_data(data_dir, benchmark_tag="M2a")
plotter.plot_volumetric_strain_vs_angle(
vtu_files_dict_full,
model_type="M2a",
ylim_range=[-7.5, 2.5],
layout="subplots",
external_data=external_data["strain"],
)[0] info: Reading meshes took 0.00961756200000008 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9790000000075025e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.1639999999999446e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011138840000000094 s
[0] info: Reset nodes in regular elements took 0.005195585999999919 s
[0] info: creation of merged mesh took 0.00047063199999985983 s
[0] info: merge properties into merged mesh took 0.0003394530000000451 s
[0] info: writing mesh took 0.012139214999999925 s
[0] info: Reading meshes took 0.012067266999999937 s
[0] info: Collection of 2921 regular elements and computing element map took 1.994999999999081e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.165000000082685e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011162980000001266 s
[0] info: Reset nodes in regular elements took 0.005357571000000005 s
[0] info: creation of merged mesh took 0.0004735559999999861 s
[0] info: merge properties into merged mesh took 0.00034171699999996363 s
[0] info: writing mesh took 0.013004534000000012 s
[0] info: Reading meshes took 0.014264789000000055 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9740000000156854e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 2.995000000005632e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011180500000000926 s
[0] info: Reset nodes in regular elements took 0.005246513000000119 s
[0] info: creation of merged mesh took 0.0004618679999999209 s
[0] info: merge properties into merged mesh took 0.0003500489999999079 s
[0] info: writing mesh took 0.012233998000000135 s
[0] info: Reading meshes took 0.009960772000000118 s
[0] info: Collection of 2921 regular elements and computing element map took 1.975999999981326e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.0450000001458477e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011153360000000223 s
[0] info: Reset nodes in regular elements took 0.0052449009999999685 s
[0] info: creation of merged mesh took 0.00046293000000008355 s
[0] info: merge properties into merged mesh took 0.0003363779999998595 s
[0] info: writing mesh took 0.011816296999999976 s
[0] info: Reading meshes took 0.00831262899999996 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9248999999721406e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 2.8349999998678044e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011151059999998658 s
[0] info: Reset nodes in regular elements took 0.005263538999999984 s
[0] info: creation of merged mesh took 0.00046567400000019354 s
[0] info: merge properties into merged mesh took 0.0003510410000000519 s
[0] info: writing mesh took 0.012553883000000043 s
[0] info: Reading meshes took 0.006839632000000151 s
[0] info: Collection of 2921 regular elements and computing element map took 1.209800000001593e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 2.013000000022913e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0006920559999998854 s
[0] info: Reset nodes in regular elements took 0.003228751999999835 s
[0] info: creation of merged mesh took 0.00029111000000003884 s
[0] info: merge properties into merged mesh took 0.00021739900000006251 s
[0] info: writing mesh took 0.009038426000000044 s

plotter.plot_field_variables(vtu_files_dict_full)Load: A, File 0, Benchmark: M2a, Material: Gneiss
[0] info: Reading meshes took 0.008998776999999958 s
[0] info: Collection of 2921 regular elements and computing element map took 2.0901999999933807e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.5560000000156577e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.001197620999999982 s
[0] info: Reset nodes in regular elements took 0.005637223999999996 s
[0] info: creation of merged mesh took 0.000495609000000119 s
[0] info: merge properties into merged mesh took 0.0003661339999998514 s
[0] info: writing mesh took 0.013689288999999993 s

Load: A, File 1, Benchmark: M2a, Material: Greywacke
[0] info: Reading meshes took 0.01130100799999978 s
[0] info: Collection of 2921 regular elements and computing element map took 2.075199999995725e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.2350000001013512e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0013590050000000797 s
[0] info: Reset nodes in regular elements took 0.005608430000000109 s
[0] info: creation of merged mesh took 0.0004947379999999946 s
[0] info: merge properties into merged mesh took 0.00036759600000002557 s
[0] info: writing mesh took 0.013580425000000007 s

Load: B, File 0, Benchmark: M2a, Material: Gneiss
[0] info: Reading meshes took 0.012843751000000125 s
[0] info: Collection of 2921 regular elements and computing element map took 1.955999999991853e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.3750000001386837e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.001214056000000019 s
[0] info: Reset nodes in regular elements took 0.005204108999999901 s
[0] info: creation of merged mesh took 0.0004804659999999572 s
[0] info: merge properties into merged mesh took 0.00034028500000005124 s
[0] info: writing mesh took 0.013563849999999933 s

Load: B, File 1, Benchmark: M2a, Material: Greywacke
[0] info: Reading meshes took 0.012605871000000102 s
[0] info: Collection of 2921 regular elements and computing element map took 2.877400000000918e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 4.547000000076906e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0014247950000001453 s
[0] info: Reset nodes in regular elements took 0.00559213599999997 s
[0] info: creation of merged mesh took 0.0004920950000000257 s
[0] info: merge properties into merged mesh took 0.000530692999999971 s
[0] info: writing mesh took 0.013545591999999829 s

Load: C, File 0, Benchmark: M2a, Material: Gneiss
[0] info: Reading meshes took 0.010971618999999988 s
[0] info: Collection of 2921 regular elements and computing element map took 2.031999999996259e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.1350000000429645e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.001287256999999986 s
[0] info: Reset nodes in regular elements took 0.005608701000000105 s
[0] info: creation of merged mesh took 0.000513256000000073 s
[0] info: merge properties into merged mesh took 0.00037859299999998264 s
[0] info: writing mesh took 0.014127021000000184 s

Load: C, File 1, Benchmark: M2a, Material: Greywacke
[0] info: Reading meshes took 0.011740863000000212 s
[0] info: Collection of 2921 regular elements and computing element map took 2.154300000012377e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.254999999979802e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0012103699999999051 s
[0] info: Reset nodes in regular elements took 0.005694781000000093 s
[0] info: creation of merged mesh took 0.0005085689999999587 s
[0] info: merge properties into merged mesh took 0.0003694989999998288 s
[0] info: writing mesh took 0.014618845000000213 s

Half fractured sample with fluid injection ($\texttt{HM}_{2b}$)
We use all inputs and BCs from $\texttt{M}_{2b}$ for a single wing fracture $\Gamma = [0,\,0.04]\times\{0\}.$ A Dirichlet BC of $p(0.04,0)=3.45\text{ MPa}$ is applied at the fracture’s right tip. After a 3000 s equilibration, fluid is injected at the left tip with $ Q_0^{\mathrm{v}}(0,0)=4.167\times10^{-7}\,\mathrm{m}^3/\mathrm{s}\;(25\text{ ml/min}).$
Run the simulation
Inputs
# Times for load curves…
(click to toggle)
# Times for load curves
times = "0.0 1000. 3500"
simulation_end_time = 3500.0
n_fracture_p_ncs = 3
model_type = "HM2b"
output_prefix = "HM2b_VPF"
# Load
PEE_load_values = {
"A": [10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6, 9.95e6],
"E": [9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6],
"B": [7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6],
"F": [3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6],
"C": [1.0e6, 3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6],
}
for p in materials.values():
p["fluid"][
"injectionFlowRate_Inlet"
] = 4.167e-7 # Injection flow rate m³/s (25 ml/min)
# Project file
prj_file = Path("HM2b_VPF.prj")
prj = ot.Project(input_file=prj_file, output_file=Path(out_dir, f"{output_prefix}.prj"))Run OGS
sing_ogs_model = SingleOGSModel(…
(click to toggle)
sing_ogs_model = SingleOGSModel(
model=prj,
out_dir=out_dir,
mesh_path=mesh_path_VPF,
output_prefix=output_prefix,
method="VPF",
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
materials=materials,
n_mpi=2,
)
vtu_files_dict_embedded_HM = sing_ogs_model.run_simulations_with_fracture(
times=times,
base_project_file=prj_file,
mesh_path=mesh_path_VPF,
load_cases=PEE_load_values,
material_names=material_names,
materials=materials,
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
output_prefix=output_prefix,
out_dir=out_dir,
method="VPF",
crack_type="half",
fracture_model_type="Isotropic",
mesh_size=h,
n_mpi=2,
)mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF.prj
Modifying mesh for VPF method at /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF with crack type: half and h1: 0.001
[0] info: Mesh 'domain' read: 2785 nodes, 2921 elements.
[0] info: Property global_node_ids is added to mesh domain
[0] info: Property global_element_ids is added to mesh domain
[0] info: Write the mesh into METIS input file.
[0] info: Total runtime: 0.0126134 s.
[0] info: Total CPU time: 0.012457 s.
[0] info: Mesh 'domain' read: 2785 nodes, 2921 elements.
[0] info: Property global_node_ids is added to mesh domain
[0] info: Property global_element_ids is added to mesh domain
[0] info: METIS is running ...
[0] info: Path to mpmetis is:
[0] info: Running: mpmetis -gtype=nodal "/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/domain.mesh" 2
******************************************************************************
METIS 5.2.1 Copyright 1998-22, Regents of the University of Minnesota
(HEAD: unknown, Built on: Sep 26 2025, 11:30:11)
size of idx_t: 64bits, real_t: 32bits, idx_t *: 64bits
Mesh Information ------------------------------------------------------------
Name: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/domain.mesh, #Elements: 2921, #Nodes: 2785, #Parts: 2
Options ---------------------------------------------------------------------
ptype=kway, objtype=cut, ctype=shem, rtype=greedy, iptype=metisrb
dbglvl=0, ufactor=1.030, minconn=NO, contig=NO, nooutput=NO
seed=-1, niter=10, ncuts=1
gtype=nodal, ncommon=1, niter=10, ncuts=1
Direct k-way Partitioning ---------------------------------------------------
- Edgecut: 119.
Timing Information ----------------------------------------------------------
I/O: 0.001 sec
Partitioning: 0.002 sec (METIS time)
Reporting: 0.000 sec
Memory Information ----------------------------------------------------------
Max memory used: 0.902 MB
rusage.ru_maxrss: 108.793 MB
proc/self/stat/VmPeak: 25.488 MB
******************************************************************************
[0] info: Partitioning the mesh in the node wise way ...
[0] info: partitionMesh(): Partition IDs per element computed in 0.000129967 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.4702e-05 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 0.000177569 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 0.000110697 s
[0] info: partitionMesh(): distribute elements into partitions took 8.9516e-05 s
[0] info: partitionMesh(): determine / append ghost nodes took 1.947e-05 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.042e-06 s
[0] info: Partitioning other meshes according to the main mesh partitions.
[0] info: Mesh 'physical_group_DSS1' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS1.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS1
[0] info: Property global_element_ids is added to mesh physical_group_DSS1
[0] info: partitionMesh(): Partition IDs per element computed in 2.6e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.9e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.31e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.91e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.5e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.21e-07 s
[0] info: Mesh 'physical_group_DSS1a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS1a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS1a
[0] info: Property global_element_ids is added to mesh physical_group_DSS1a
[0] info: partitionMesh(): Partition IDs per element computed in 2.6e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.1e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.51e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.8e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.81e-07 s
[0] info: Mesh 'physical_group_DSS2' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS2.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS2
[0] info: Property global_element_ids is added to mesh physical_group_DSS2
[0] info: partitionMesh(): Partition IDs per element computed in 2.7e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.71e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 4.7e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.6e-07 s
[0] info: Mesh 'physical_group_DSS2a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS2a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS2a
[0] info: Property global_element_ids is added to mesh physical_group_DSS2a
[0] info: partitionMesh(): Partition IDs per element computed in 2.5e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.81e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.21e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.7e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.6e-07 s
[0] info: Mesh 'physical_group_DSS3' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS3.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS3
[0] info: Property global_element_ids is added to mesh physical_group_DSS3
[0] info: partitionMesh(): Partition IDs per element computed in 3.1e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.21e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.71e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.4e-07 s
[0] info: Mesh 'physical_group_DSS3a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS3a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS3a
[0] info: Property global_element_ids is added to mesh physical_group_DSS3a
[0] info: partitionMesh(): Partition IDs per element computed in 2.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.9e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.5e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.41e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.6e-07 s
[0] info: Mesh 'physical_group_DSS4' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS4.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS4
[0] info: Property global_element_ids is added to mesh physical_group_DSS4
[0] info: partitionMesh(): Partition IDs per element computed in 2.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.9e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.4e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.4e-07 s
[0] info: Mesh 'physical_group_DSS4a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS4a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS4a
[0] info: Property global_element_ids is added to mesh physical_group_DSS4a
[0] info: partitionMesh(): Partition IDs per element computed in 2.31e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.1e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.31e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.7e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.91e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.5e-07 s
[0] info: Mesh 'physical_group_DSS5' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS5.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS5
[0] info: Property global_element_ids is added to mesh physical_group_DSS5
[0] info: partitionMesh(): Partition IDs per element computed in 2.61e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.11e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.8e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.71e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.6e-07 s
[0] info: Mesh 'physical_group_DSS5a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS5a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS5a
[0] info: Property global_element_ids is added to mesh physical_group_DSS5a
[0] info: partitionMesh(): Partition IDs per element computed in 2.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.21e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.4e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.3e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.9e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.71e-07 s
[0] info: Mesh 'physical_group_DSS6' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS6.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS6
[0] info: Property global_element_ids is added to mesh physical_group_DSS6
[0] info: partitionMesh(): Partition IDs per element computed in 2.31e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.8e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.3e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.6e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.71e-07 s
[0] info: Mesh 'physical_group_DSS6a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS6a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS6a
[0] info: Property global_element_ids is added to mesh physical_group_DSS6a
[0] info: partitionMesh(): Partition IDs per element computed in 2.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.81e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.5e-07 s
[0] info: Mesh 'physical_group_DSS7' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS7.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS7
[0] info: Property global_element_ids is added to mesh physical_group_DSS7
[0] info: partitionMesh(): Partition IDs per element computed in 2.51e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.4e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.4e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_DSS7a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS7a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS7a
[0] info: Property global_element_ids is added to mesh physical_group_DSS7a
[0] info: partitionMesh(): Partition IDs per element computed in 2.5e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.3e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.5e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.41e-07 s
[0] info: Mesh 'physical_group_DSS8' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS8.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS8
[0] info: Property global_element_ids is added to mesh physical_group_DSS8
[0] info: partitionMesh(): Partition IDs per element computed in 2.5e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.1e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3.4e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.4e-07 s
[0] info: Mesh 'physical_group_DSS8a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS8a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS8a
[0] info: Property global_element_ids is added to mesh physical_group_DSS8a
[0] info: partitionMesh(): Partition IDs per element computed in 2.6e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 4.21e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.41e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.21e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.6e-07 s
[0] info: Mesh 'physical_group_PEE1' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE1.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE1
[0] info: Property global_element_ids is added to mesh physical_group_PEE1
[0] info: partitionMesh(): Partition IDs per element computed in 3.5e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.81e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.61e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.4e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.71e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.4e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.4e-07 s
[0] info: Mesh 'physical_group_PEE1a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE1a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE1a
[0] info: Property global_element_ids is added to mesh physical_group_PEE1a
[0] info: partitionMesh(): Partition IDs per element computed in 3.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.71e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.61e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.41e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.4e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.4e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.5e-07 s
[0] info: Mesh 'physical_group_PEE2' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE2.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE2
[0] info: Property global_element_ids is added to mesh physical_group_PEE2
[0] info: partitionMesh(): Partition IDs per element computed in 4.5e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.3e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.8e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.5e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.4e-07 s
[0] info: Mesh 'physical_group_PEE2a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE2a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE2a
[0] info: Property global_element_ids is added to mesh physical_group_PEE2a
[0] info: partitionMesh(): Partition IDs per element computed in 3.01e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.1e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.41e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.3e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.71e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.31e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.3e-07 s
[0] info: Mesh 'physical_group_PEE3' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE3.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE3
[0] info: Property global_element_ids is added to mesh physical_group_PEE3
[0] info: partitionMesh(): Partition IDs per element computed in 3.2e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.31e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.6e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_PEE3a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE3a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE3a
[0] info: Property global_element_ids is added to mesh physical_group_PEE3a
[0] info: partitionMesh(): Partition IDs per element computed in 3.91e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.81e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.61e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.71e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.5e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.6e-07 s
[0] info: Mesh 'physical_group_PEE4' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE4.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE4
[0] info: Property global_element_ids is added to mesh physical_group_PEE4
[0] info: partitionMesh(): Partition IDs per element computed in 2.81e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.3e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.5e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.8e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.4e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.61e-07 s
[0] info: Mesh 'physical_group_PEE4a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE4a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE4a
[0] info: Property global_element_ids is added to mesh physical_group_PEE4a
[0] info: partitionMesh(): Partition IDs per element computed in 3.2e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 5.71e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 7.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.21e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 6.01e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 8.41e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2.9e-07 s
[0] info: Mesh 'physical_group_PEE5' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE5.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE5
[0] info: Property global_element_ids is added to mesh physical_group_PEE5
[0] info: partitionMesh(): Partition IDs per element computed in 2.91e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 4.81e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 7.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.8e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.3e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 8.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2.61e-07 s
[0] info: Mesh 'physical_group_PEE5a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE5a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE5a
[0] info: Property global_element_ids is added to mesh physical_group_PEE5a
[0] info: partitionMesh(): Partition IDs per element computed in 3.21e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.61e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 7.01e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.9e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.5e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.6e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.4e-07 s
[0] info: Mesh 'physical_group_PEE6' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE6.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE6
[0] info: Property global_element_ids is added to mesh physical_group_PEE6
[0] info: partitionMesh(): Partition IDs per element computed in 2.8e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.61e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.61e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.31e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.61e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.6e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.4e-07 s
[0] info: Mesh 'physical_group_PEE6a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE6a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE6a
[0] info: Property global_element_ids is added to mesh physical_group_PEE6a
[0] info: partitionMesh(): Partition IDs per element computed in 3.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.6e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.31e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.31e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.61e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.9e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.3e-07 s
[0] info: Mesh 'physical_group_PEE7' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE7.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE7
[0] info: Property global_element_ids is added to mesh physical_group_PEE7
[0] info: partitionMesh(): Partition IDs per element computed in 3.11e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.21e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.5e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.71e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_PEE7a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE7a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE7a
[0] info: Property global_element_ids is added to mesh physical_group_PEE7a
[0] info: partitionMesh(): Partition IDs per element computed in 3.2e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.1e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.6e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.6e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.61e-07 s
[0] info: Mesh 'physical_group_PEE8' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE8.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE8
[0] info: Property global_element_ids is added to mesh physical_group_PEE8
[0] info: partitionMesh(): Partition IDs per element computed in 2.9e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.31e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.51e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.51e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.8e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.4e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.41e-07 s
[0] info: Mesh 'physical_group_PEE8a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE8a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE8a
[0] info: Property global_element_ids is added to mesh physical_group_PEE8a
[0] info: partitionMesh(): Partition IDs per element computed in 2.6e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.51e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.31e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.3e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.41e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.4e-07 s
[0] info: Mesh 'physical_group_p_bottom' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_bottom.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_bottom
[0] info: Property global_element_ids is added to mesh physical_group_p_bottom
[0] info: partitionMesh(): Partition IDs per element computed in 2.3e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 8e-08 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.3e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 8e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2.11e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.5e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_p_left' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_left.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_left
[0] info: Property global_element_ids is added to mesh physical_group_p_left
[0] info: partitionMesh(): Partition IDs per element computed in 2.11e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 8e-08 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 8e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 1.7e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.81e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.61e-07 s
[0] info: Mesh 'physical_group_p_right' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_right.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_right
[0] info: Property global_element_ids is added to mesh physical_group_p_right
[0] info: partitionMesh(): Partition IDs per element computed in 2e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 7e-08 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 7.1e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 1.8e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.41e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.6e-07 s
[0] info: Mesh 'physical_group_p_top' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_top.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_top
[0] info: Property global_element_ids is added to mesh physical_group_p_top
[0] info: partitionMesh(): Partition IDs per element computed in 2e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 8e-08 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 8e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.6e-07 s
[0] info: Mesh 'physical_group_Inlet' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Inlet.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Inlet
[0] info: Property global_element_ids is added to mesh physical_group_Inlet
[0] info: partitionMesh(): Partition IDs per element computed in 1.8e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.7e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 1.9e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.6e-07 s
[0] info: Mesh 'physical_group_Outlet_R_embeddedFracture' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_R_embeddedFracture.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Outlet_R_embeddedFracture
[0] info: Property global_element_ids is added to mesh physical_group_Outlet_R_embeddedFracture
[0] info: partitionMesh(): Partition IDs per element computed in 1.9e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.71e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.01e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 2e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.71e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.4e-07 s
[0] info: Mesh 'physical_group_Outlet_R_fullFracture' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_R_fullFracture.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Outlet_R_fullFracture
[0] info: Property global_element_ids is added to mesh physical_group_Outlet_R_fullFracture
[0] info: partitionMesh(): Partition IDs per element computed in 2.21e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.3e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.01e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 2e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.41e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.61e-07 s
[0] info: Mesh 'physical_group_Outlet_L_fullFracture' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_L_fullFracture.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Outlet_L_fullFracture
[0] info: Property global_element_ids is added to mesh physical_group_Outlet_L_fullFracture
[0] info: partitionMesh(): Partition IDs per element computed in 1.9e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.3e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 2.01e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.6e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.6e-07 s
[0] info: Writing the partitions data into binary files took 0.00152335 s
[0] info: Total runtime: 0.103394 s.
[0] info: Total CPU time: 0.054315 s.
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_A.prj
============================================================
Running simulation for load case: A with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_A.prj
Status: finished successfully.
Execution took 41.86760592460632 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_A.prj
Status: finished successfully.
Execution took 50.784921646118164 s
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_E.prj
============================================================
Running simulation for load case: E with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_E.prj
Status: finished successfully.
Execution took 41.40125799179077 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_E.prj
Status: finished successfully.
Execution took 50.445422410964966 s
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_B.prj
============================================================
Running simulation for load case: B with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_B.prj
Status: finished successfully.
Execution took 41.008578062057495 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_B.prj
Status: finished successfully.
Execution took 50.40303182601929 s
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_F.prj
============================================================
Running simulation for load case: F with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_F.prj
Status: finished successfully.
Execution took 40.952221632003784 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_F.prj
Status: finished successfully.
Execution took 50.252476930618286 s
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_C.prj
============================================================
Running simulation for load case: C with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_C.prj
Status: finished successfully.
Execution took 40.631916761398315 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2b_VPF_C.prj
Status: finished successfully.
Execution took 55.75521206855774 s
Post-processing
plotter = Plotter(…
(click to toggle)
plotter = Plotter(
output_dir=out_dir,
save_extracted_data=True,
)
data_dir = Path("external_data")
external_data = Plotter.load_external_data(data_dir, benchmark_tag="HM2b")
plotter.plot_volumetric_strain_vs_angle(
vtu_files_dict_embedded_HM,
model_type="HM2b",
ylim_range=[-7.5, 2.5],
layout="subplots",
external_data=external_data["strain"],
)[0] info: Reading meshes took 0.01233332999999992 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9589999999958252e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.3849999998558644e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011180900000000715 s
[0] info: Reset nodes in regular elements took 0.005332883999999982 s
[0] info: creation of merged mesh took 0.0004587929999999574 s
[0] info: merge properties into merged mesh took 0.0003583120000001383 s
[0] info: writing mesh took 0.01077262999999995 s
[0] info: Reading meshes took 0.012839274999999928 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9208999999964504e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 2.9939999999228917e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.001123779000000047 s
[0] info: Reset nodes in regular elements took 0.005333255000000037 s
[0] info: creation of merged mesh took 0.0004740970000001177 s
[0] info: merge properties into merged mesh took 0.00034612299999992047 s
[0] info: writing mesh took 0.01269426600000001 s
[0] info: Reading meshes took 0.012035249000000192 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9760000000035305e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.044999999923803e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011262830000000612 s
[0] info: Reset nodes in regular elements took 0.00533842200000012 s
[0] info: creation of merged mesh took 0.0004554580000000197 s
[0] info: merge properties into merged mesh took 0.00035382500000014083 s
[0] info: writing mesh took 0.012774106000000174 s
[0] info: Reading meshes took 0.010039219999999904 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9109000000128162e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.0139999998013423e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011182810000001098 s
[0] info: Reset nodes in regular elements took 0.005189477000000053 s
[0] info: creation of merged mesh took 0.0005766829999998002 s
[0] info: merge properties into merged mesh took 0.0003428180000000225 s
[0] info: writing mesh took 0.01254379800000005 s
[0] info: Reading meshes took 0.010369781000000078 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9239000000004225e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.124000000020999e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011180910000001543 s
[0] info: Reset nodes in regular elements took 0.005321756999999927 s
[0] info: creation of merged mesh took 0.00046635499999991836 s
[0] info: merge properties into merged mesh took 0.00035045000000000215 s
[0] info: writing mesh took 0.012443447000000107 s
[0] info: Reading meshes took 0.013422026000000198 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9048000000188026e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.3150000000592428e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011226370000001484 s
[0] info: Reset nodes in regular elements took 0.0052920429999998575 s
[0] info: creation of merged mesh took 0.000468397999999981 s
[0] info: merge properties into merged mesh took 0.00035593799999977804 s
[0] info: writing mesh took 0.01005193999999987 s
[0] info: Reading meshes took 0.014084817999999943 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9589999999958252e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.2349999998793066e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.001213845999999963 s
[0] info: Reset nodes in regular elements took 0.0052200829999997644 s
[0] info: creation of merged mesh took 0.00048706600000003597 s
[0] info: merge properties into merged mesh took 0.0003551969999999738 s
[0] info: writing mesh took 0.012796501000000182 s
[0] info: Reading meshes took 0.012092857000000068 s
[0] info: Collection of 2921 regular elements and computing element map took 2.0269999999822375e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.0839999998200085e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011242200000001201 s
[0] info: Reset nodes in regular elements took 0.005321386999999955 s
[0] info: creation of merged mesh took 0.0004820690000000294 s
[0] info: merge properties into merged mesh took 0.0003479760000000276 s
[0] info: writing mesh took 0.013240361999999894 s
[0] info: Reading meshes took 0.013909514000000067 s
[0] info: Collection of 2921 regular elements and computing element map took 1.984000000021524e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.014000000023387e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011195729999999848 s
[0] info: Reset nodes in regular elements took 0.0052052809999998395 s
[0] info: creation of merged mesh took 0.00048237899999992173 s
[0] info: merge properties into merged mesh took 0.00034769500000009224 s
[0] info: writing mesh took 0.012396065000000123 s
[0] info: Reading meshes took 0.012985845999999857 s
[0] info: Collection of 2921 regular elements and computing element map took 2.5688000000023692e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.0749999999635236e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.001117519999999983 s
[0] info: Reset nodes in regular elements took 0.005303480000000027 s
[0] info: creation of merged mesh took 0.0004683090000001666 s
[0] info: merge properties into merged mesh took 0.0003518920000000758 s
[0] info: writing mesh took 0.012367971999999838 s

plotter.plot_field_variables(vtu_files_dict_embedded_HM)Load: A, File 0, Benchmark: HM2b, Material: Gneiss
[0] info: Reading meshes took 0.012590309000000133 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9599999999897477e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.1049999997811994e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011196730000000432 s
[0] info: Reset nodes in regular elements took 0.005202868000000027 s
[0] info: creation of merged mesh took 0.0004731760000000751 s
[0] info: merge properties into merged mesh took 0.0003532340000000911 s
[0] info: writing mesh took 0.013093650000000068 s

Load: A, File 1, Benchmark: HM2b, Material: Greywacke
[0] info: Reading meshes took 0.013242935000000067 s
[0] info: Collection of 2921 regular elements and computing element map took 2.007000000014969e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.2440000001798808e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011220170000001417 s
[0] info: Reset nodes in regular elements took 0.005306274000000055 s
[0] info: creation of merged mesh took 0.00047035099999992447 s
[0] info: merge properties into merged mesh took 0.0003505300000001821 s
[0] info: writing mesh took 0.014735381999999797 s

Load: E, File 0, Benchmark: HM2b, Material: Gneiss
[0] info: Reading meshes took 0.011967926999999934 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9699999999955864e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.254999999979802e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.001121685999999844 s
[0] info: Reset nodes in regular elements took 0.005326564999999839 s
[0] info: creation of merged mesh took 0.000466856000000071 s
[0] info: merge properties into merged mesh took 0.00034730500000001996 s
[0] info: writing mesh took 0.013194533000000064 s

Load: E, File 1, Benchmark: HM2b, Material: Greywacke
[0] info: Reading meshes took 0.00944992 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9580000000019027e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.304999999897973e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011187420000000614 s
[0] info: Reset nodes in regular elements took 0.0052061029999999064 s
[0] info: creation of merged mesh took 0.0004700609999999106 s
[0] info: merge properties into merged mesh took 0.00034743599999997876 s
[0] info: writing mesh took 0.012774126999999913 s

Load: B, File 0, Benchmark: HM2b, Material: Gneiss
[0] info: Reading meshes took 0.00829502400000015 s
[0] info: Collection of 2921 regular elements and computing element map took 1.954000000004008e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.0649999998022537e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011196930000001437 s
[0] info: Reset nodes in regular elements took 0.005283459000000157 s
[0] info: creation of merged mesh took 0.000464883000000027 s
[0] info: merge properties into merged mesh took 0.0003439000000000636 s
[0] info: writing mesh took 0.013311669000000137 s

Load: B, File 1, Benchmark: HM2b, Material: Greywacke
[0] info: Reading meshes took 0.014235145000000005 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9500000000061135e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.2550000002018464e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011190819999999935 s
[0] info: Reset nodes in regular elements took 0.005211730999999942 s
[0] info: creation of merged mesh took 0.0004712329999998488 s
[0] info: merge properties into merged mesh took 0.00034404100000018367 s
[0] info: writing mesh took 0.013261262999999968 s

Load: F, File 0, Benchmark: HM2b, Material: Gneiss
[0] info: Reading meshes took 0.014281596000000008 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9619999999997972e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.1249999998816946e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011248410000002096 s
[0] info: Reset nodes in regular elements took 0.0051905590000000945 s
[0] info: creation of merged mesh took 0.0004788039999998883 s
[0] info: merge properties into merged mesh took 0.000351521999999882 s
[0] info: writing mesh took 0.013112388000000141 s

Load: F, File 1, Benchmark: HM2b, Material: Greywacke
[0] info: Reading meshes took 0.01171248200000008 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9680000000077413e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.1249999998816946e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011158470000001142 s
[0] info: Reset nodes in regular elements took 0.00531575899999992 s
[0] info: creation of merged mesh took 0.0004814280000000615 s
[0] info: merge properties into merged mesh took 0.00034647400000009654 s
[0] info: writing mesh took 0.013225209000000016 s

Load: C, File 0, Benchmark: HM2b, Material: Gneiss
[0] info: Reading meshes took 0.010032751000000006 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9839999999993196e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.034000000123882e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011288469999999329 s
[0] info: Reset nodes in regular elements took 0.005202797000000148 s
[0] info: creation of merged mesh took 0.0004677169999998121 s
[0] info: merge properties into merged mesh took 0.000438652999999789 s
[0] info: writing mesh took 0.012914027999999966 s

Load: C, File 1, Benchmark: HM2b, Material: Greywacke
[0] info: Reading meshes took 0.012010100999999995 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9849999999932422e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.3150000000592428e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011154070000001237 s
[0] info: Reset nodes in regular elements took 0.00531183199999985 s
[0] info: creation of merged mesh took 0.00047171300000004024 s
[0] info: merge properties into merged mesh took 0.00034709499999996396 s
[0] info: writing mesh took 0.013073839000000032 s

plotter.material_names = ["Gneiss", "Greywacke"]…
(click to toggle)
plotter.material_names = ["Gneiss", "Greywacke"]
plotter.vtu_file_names = {"VPF": vtu_files_dict_embedded_HM}
plotter.plot_avg_width_vs_stress(
benchmark_tag="HM2b",
metric="width",
methods_to_include=["VPF"],
pee_load_values=PEE_load_values,
external_data=external_data["average"],
ylim_range=(0, 10e-6),
)
plotter.plot_avg_width_vs_stress(
benchmark_tag="HM2b",
metric="permeability",
methods_to_include=["VPF"],
pee_load_values=PEE_load_values,
external_data=external_data["average"],
ylim_range=(0, 5e-12),
)


array([ 1000000. , 2318019.48466054, 5500000. ,
8681980.51533946, 10000000. ])
vpf_profiles = {}…
(click to toggle)
vpf_profiles = {}
for load_case, file_list in plotter.vtu_file_names["VPF"].items():
vpf_profiles[load_case] = plotter.extract_vpf_width_from_list(
file_list,
plotter.material_names,
)
plotter.plot_fracture_aperture_profiles(
widthProfile=vpf_profiles,
benchmark_tag="HM2b",
downsample=1,
ylim=(0, 10e-6),
method_label="VPF",
external_data=external_data["widthProfile"],
)





Fully fractured sample with fluid injection ($\texttt{HM}_{2a}$)
We performed five hydro-mechanical (HMca) simulations with different angles between the second principal stress direction (σ₂ = 10 MPa) and the PEE 5 fracture plane $\Gamma = [-0.094,\,0.094]\times\{0\}.$ The initial pore pressure was uniform at $p_0 = 0.1$ MPa, with Dirichlet conditions $p(-0.094,0)=p(0.094,0)=3.45\text{ MPa}.$ After equilibrating for 3000 s, we injected fluid at the center at a rate $ Q_0^{\text{v}}(0,0)=4.167\times10^{-7}\,\mathrm{m}^3/\mathrm{s}\;(25\,\mathrm{ml/min})$ for 500 s, starting from an initial fracture aperture $w_{\text{ini}}=1\times10^{-6}$ m.
Run the simulation
Inputs
# Times for load curves…
(click to toggle)
# Times for load curves
times = "0.0 1000. 3500"
simulation_end_time = 3500.0
n_fracture_p_ncs = 3
model_type = "HM2a"
output_prefix = "HM2a_VPF"
# Load
PEE_load_values = {
"A": [10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6, 9.95e6],
# "E": [9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6],
# "B": [7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6],
# "F": [3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6],
# "C": [1.0e6, 3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6],
}
# To save time, we only run the first load case A.
# If you want to run all load cases, uncomment the lines above.
for p in materials.values():
p["fluid"][
"injectionFlowRate_Inlet"
] = 4.167e-7 # Injection flow rate m³/s (25 ml/min)
# Project file
prj_file = Path("HM2a_VPF.prj")
prj = ot.Project(input_file=prj_file, output_file=Path(out_dir, f"{output_prefix}.prj"))Run OGS
sing_ogs_model = SingleOGSModel(…
(click to toggle)
sing_ogs_model = SingleOGSModel(
model=prj,
out_dir=out_dir,
mesh_path=mesh_path_VPF,
output_prefix=output_prefix,
method="VPF",
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
materials=materials,
n_mpi=2,
)
vtu_files_dict_full_HM = sing_ogs_model.run_simulations_with_fracture(
times=times,
base_project_file=prj_file,
mesh_path=mesh_path_VPF,
load_cases=PEE_load_values,
material_names=material_names,
materials=materials,
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
output_prefix=output_prefix,
out_dir=out_dir,
method="VPF",
crack_type="full",
fracture_model_type="Isotropic",
mesh_size=h,
n_mpi=2,
)mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2a_VPF.prj
Modifying mesh for VPF method at /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF with crack type: full and h1: 0.001
[0] info: Mesh 'domain' read: 2785 nodes, 2921 elements.
[0] info: Property global_node_ids is added to mesh domain
[0] info: Property global_element_ids is added to mesh domain
[0] info: Write the mesh into METIS input file.
[0] info: Total runtime: 0.00964623 s.
[0] info: Total CPU time: 0.009565 s.
[0] info: Mesh 'domain' read: 2785 nodes, 2921 elements.
[0] info: Property global_node_ids is added to mesh domain
[0] info: Property global_element_ids is added to mesh domain
[0] info: METIS is running ...
[0] info: Path to mpmetis is:
[0] info: Running: mpmetis -gtype=nodal "/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/domain.mesh" 2
******************************************************************************
METIS 5.2.1 Copyright 1998-22, Regents of the University of Minnesota
(HEAD: unknown, Built on: Sep 26 2025, 11:30:11)
size of idx_t: 64bits, real_t: 32bits, idx_t *: 64bits
Mesh Information ------------------------------------------------------------
Name: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/domain.mesh, #Elements: 2921, #Nodes: 2785, #Parts: 2
Options ---------------------------------------------------------------------
ptype=kway, objtype=cut, ctype=shem, rtype=greedy, iptype=metisrb
dbglvl=0, ufactor=1.030, minconn=NO, contig=NO, nooutput=NO
seed=-1, niter=10, ncuts=1
gtype=nodal, ncommon=1, niter=10, ncuts=1
Direct k-way Partitioning ---------------------------------------------------
- Edgecut: 119.
Timing Information ----------------------------------------------------------
I/O: 0.001 sec
Partitioning: 0.002 sec (METIS time)
Reporting: -0.000 sec
Memory Information ----------------------------------------------------------
Max memory used: 0.902 MB
rusage.ru_maxrss: 109.230 MB
proc/self/stat/VmPeak: 25.488 MB
******************************************************************************
[0] info: Partitioning the mesh in the node wise way ...
[0] info: partitionMesh(): Partition IDs per element computed in 0.000174604 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.0892e-05 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 0.000274725 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 0.000190939 s
[0] info: partitionMesh(): distribute elements into partitions took 0.000126832 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.1157e-05 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.252e-06 s
[0] info: Partitioning other meshes according to the main mesh partitions.
[0] info: Mesh 'physical_group_DSS1' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS1.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS1
[0] info: Property global_element_ids is added to mesh physical_group_DSS1
[0] info: partitionMesh(): Partition IDs per element computed in 3.3e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.21e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 8.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 2.31e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 6.11e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 6.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2.7e-07 s
[0] info: Mesh 'physical_group_DSS1a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS1a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS1a
[0] info: Property global_element_ids is added to mesh physical_group_DSS1a
[0] info: partitionMesh(): Partition IDs per element computed in 4.5e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.71e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 9.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 2.4e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 6.21e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 6.21e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.21e-07 s
[0] info: Mesh 'physical_group_DSS2' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS2.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS2
[0] info: Property global_element_ids is added to mesh physical_group_DSS2
[0] info: partitionMesh(): Partition IDs per element computed in 3.1e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.61e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 8.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 2.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.81e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.6e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.2e-07 s
[0] info: Mesh 'physical_group_DSS2a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS2a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS2a
[0] info: Property global_element_ids is added to mesh physical_group_DSS2a
[0] info: partitionMesh(): Partition IDs per element computed in 3.21e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.11e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 9.41e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.91e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3e-07 s
[0] info: Mesh 'physical_group_DSS3' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS3.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS3
[0] info: Property global_element_ids is added to mesh physical_group_DSS3
[0] info: partitionMesh(): Partition IDs per element computed in 3.21e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.4e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 8.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.9e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.71e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3e-07 s
[0] info: Mesh 'physical_group_DSS3a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS3a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS3a
[0] info: Property global_element_ids is added to mesh physical_group_DSS3a
[0] info: partitionMesh(): Partition IDs per element computed in 3.31e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.41e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 8.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 2.1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.61e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.11e-07 s
[0] info: Mesh 'physical_group_DSS4' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS4.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS4
[0] info: Property global_element_ids is added to mesh physical_group_DSS4
[0] info: partitionMesh(): Partition IDs per element computed in 3.11e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 8.82e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.8e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 6.01e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.41e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.1e-07 s
[0] info: Mesh 'physical_group_DSS4a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS4a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS4a
[0] info: Property global_element_ids is added to mesh physical_group_DSS4a
[0] info: partitionMesh(): Partition IDs per element computed in 3.2e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.7e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 9.62e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.61e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.81e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.21e-07 s
[0] info: Mesh 'physical_group_DSS5' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS5.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS5
[0] info: Property global_element_ids is added to mesh physical_group_DSS5
[0] info: partitionMesh(): Partition IDs per element computed in 3.2e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 4.21e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 8.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 2.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 6.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.11e-07 s
[0] info: Mesh 'physical_group_DSS5a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS5a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS5a
[0] info: Property global_element_ids is added to mesh physical_group_DSS5a
[0] info: partitionMesh(): Partition IDs per element computed in 3.31e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.81e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 8.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.8e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.9e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.11e-07 s
[0] info: Mesh 'physical_group_DSS6' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS6.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS6
[0] info: Property global_element_ids is added to mesh physical_group_DSS6
[0] info: partitionMesh(): Partition IDs per element computed in 3.51e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.5e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 8.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 2.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.41e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.21e-07 s
[0] info: Mesh 'physical_group_DSS6a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS6a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS6a
[0] info: Property global_element_ids is added to mesh physical_group_DSS6a
[0] info: partitionMesh(): Partition IDs per element computed in 3.81e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.5e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 8.41e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.41e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.81e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.21e-07 s
[0] info: Mesh 'physical_group_DSS7' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS7.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS7
[0] info: Property global_element_ids is added to mesh physical_group_DSS7
[0] info: partitionMesh(): Partition IDs per element computed in 3.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.3e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 8.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.71e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.61e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.31e-07 s
[0] info: Mesh 'physical_group_DSS7a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS7a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS7a
[0] info: Property global_element_ids is added to mesh physical_group_DSS7a
[0] info: partitionMesh(): Partition IDs per element computed in 3.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.21e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 8.41e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.71e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.71e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.11e-07 s
[0] info: Mesh 'physical_group_DSS8' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS8.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS8
[0] info: Property global_element_ids is added to mesh physical_group_DSS8
[0] info: partitionMesh(): Partition IDs per element computed in 3.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.3e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 9.31e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.9e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.6e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.41e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.11e-07 s
[0] info: Mesh 'physical_group_DSS8a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_DSS8a.vtu' read: 3 nodes, 2 elements.
[0] info: Property global_node_ids is added to mesh physical_group_DSS8a
[0] info: Property global_element_ids is added to mesh physical_group_DSS8a
[0] info: partitionMesh(): Partition IDs per element computed in 3.31e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.01e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 9.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 2.31e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.81e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.31e-07 s
[0] info: Mesh 'physical_group_PEE1' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE1.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE1
[0] info: Property global_element_ids is added to mesh physical_group_PEE1
[0] info: partitionMesh(): Partition IDs per element computed in 4.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 6.91e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 1.071e-06 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 2.61e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 7.61e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.91e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.1e-07 s
[0] info: Mesh 'physical_group_PEE1a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE1a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE1a
[0] info: Property global_element_ids is added to mesh physical_group_PEE1a
[0] info: partitionMesh(): Partition IDs per element computed in 5.31e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 6.01e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 9.81e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 2.3e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 7.71e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 5.81e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 3.21e-07 s
[0] info: Mesh 'physical_group_PEE2' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE2.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE2
[0] info: Property global_element_ids is added to mesh physical_group_PEE2
[0] info: partitionMesh(): Partition IDs per element computed in 3.11e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.31e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.81e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.5e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.4e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2.01e-07 s
[0] info: Mesh 'physical_group_PEE2a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE2a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE2a
[0] info: Property global_element_ids is added to mesh physical_group_PEE2a
[0] info: partitionMesh(): Partition IDs per element computed in 2.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.81e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.1e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.1e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.91e-07 s
[0] info: Mesh 'physical_group_PEE3' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE3.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE3
[0] info: Property global_element_ids is added to mesh physical_group_PEE3
[0] info: partitionMesh(): Partition IDs per element computed in 2.5e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.8e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.1e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.4e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 2.9e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_PEE3a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE3a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE3a
[0] info: Property global_element_ids is added to mesh physical_group_PEE3a
[0] info: partitionMesh(): Partition IDs per element computed in 2.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.8e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.91e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.4e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.4e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.81e-07 s
[0] info: Mesh 'physical_group_PEE4' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE4.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE4
[0] info: Property global_element_ids is added to mesh physical_group_PEE4
[0] info: partitionMesh(): Partition IDs per element computed in 2.91e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.8e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.4e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.4e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.81e-07 s
[0] info: Mesh 'physical_group_PEE4a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE4a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE4a
[0] info: Property global_element_ids is added to mesh physical_group_PEE4a
[0] info: partitionMesh(): Partition IDs per element computed in 2.91e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 4.11e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.01e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.3e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.01e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 7.21e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2.81e-07 s
[0] info: Mesh 'physical_group_PEE5' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE5.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE5
[0] info: Property global_element_ids is added to mesh physical_group_PEE5
[0] info: partitionMesh(): Partition IDs per element computed in 2.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 4.21e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.7e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.21e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 7.42e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2.8e-07 s
[0] info: Mesh 'physical_group_PEE5a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE5a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE5a
[0] info: Property global_element_ids is added to mesh physical_group_PEE5a
[0] info: partitionMesh(): Partition IDs per element computed in 2.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.41e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.81e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.5e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.3e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.91e-07 s
[0] info: Mesh 'physical_group_PEE6' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE6.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE6
[0] info: Property global_element_ids is added to mesh physical_group_PEE6
[0] info: partitionMesh(): Partition IDs per element computed in 2.41e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.4e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.4e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.2e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.11e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.41e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_PEE6a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE6a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE6a
[0] info: Property global_element_ids is added to mesh physical_group_PEE6a
[0] info: partitionMesh(): Partition IDs per element computed in 2.31e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.21e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.81e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.31e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.3e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_PEE7' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE7.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE7
[0] info: Property global_element_ids is added to mesh physical_group_PEE7
[0] info: partitionMesh(): Partition IDs per element computed in 2.3e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.01e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.61e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.3e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 5.5e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.2e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 2e-07 s
[0] info: Mesh 'physical_group_PEE7a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE7a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE7a
[0] info: Property global_element_ids is added to mesh physical_group_PEE7a
[0] info: partitionMesh(): Partition IDs per element computed in 2.3e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 6.11e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.4e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.61e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_PEE8' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE8.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE8
[0] info: Property global_element_ids is added to mesh physical_group_PEE8
[0] info: partitionMesh(): Partition IDs per element computed in 2.9e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.31e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.41e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.3e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.2e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.3e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_PEE8a' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_PEE8a.vtu' read: 5 nodes, 4 elements.
[0] info: Property global_node_ids is added to mesh physical_group_PEE8a
[0] info: Property global_element_ids is added to mesh physical_group_PEE8a
[0] info: partitionMesh(): Partition IDs per element computed in 3.61e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 3.2e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.61e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.3e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 4.51e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 4.11e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.91e-07 s
[0] info: Mesh 'physical_group_p_bottom' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_bottom.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_bottom
[0] info: Property global_element_ids is added to mesh physical_group_p_bottom
[0] info: partitionMesh(): Partition IDs per element computed in 2.8e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 8e-08 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.01e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 7e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2.2e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.11e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_p_left' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_left.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_left
[0] info: Property global_element_ids is added to mesh physical_group_p_left
[0] info: partitionMesh(): Partition IDs per element computed in 1.91e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 8e-08 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.31e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 8e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2.1e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.21e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_p_right' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_right.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_right
[0] info: Property global_element_ids is added to mesh physical_group_p_right
[0] info: partitionMesh(): Partition IDs per element computed in 1.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 9e-08 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.51e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 7e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 1.81e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.31e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_p_top' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_p_top.vtu' read: 1 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_p_top
[0] info: Property global_element_ids is added to mesh physical_group_p_top
[0] info: partitionMesh(): Partition IDs per element computed in 1.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 8e-08 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.41e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 7e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2.1e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Mesh 'physical_group_Inlet' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Inlet.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Inlet
[0] info: Property global_element_ids is added to mesh physical_group_Inlet
[0] info: partitionMesh(): Partition IDs per element computed in 2.1e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 1.51e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.9e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 2.31e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.1e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.7e-07 s
[0] info: Mesh 'physical_group_Outlet_R_embeddedFracture' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_R_embeddedFracture.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Outlet_R_embeddedFracture
[0] info: Property global_element_ids is added to mesh physical_group_Outlet_R_embeddedFracture
[0] info: partitionMesh(): Partition IDs per element computed in 1.5e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.4e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.71e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1.01e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 2.3e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.4e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.9e-07 s
[0] info: Mesh 'physical_group_Outlet_R_fullFracture' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_R_fullFracture.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Outlet_R_fullFracture
[0] info: Property global_element_ids is added to mesh physical_group_Outlet_R_fullFracture
[0] info: partitionMesh(): Partition IDs per element computed in 1.51e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.3e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 4.7e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 8e-08 s
[0] info: partitionMesh(): distribute elements into partitions took 2.3e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.6e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.81e-07 s
[0] info: Mesh 'physical_group_Outlet_L_fullFracture' from file '/var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF/physical_group_Outlet_L_fullFracture.vtu' read: 2 nodes, 1 elements.
[0] info: Property global_node_ids is added to mesh physical_group_Outlet_L_fullFracture
[0] info: Property global_element_ids is added to mesh physical_group_Outlet_L_fullFracture
[0] info: partitionMesh(): Partition IDs per element computed in 1.4e-07 s
[0] info: partitionMesh(): distribute nodes to partitions took 2.41e-07 s
[0] info: partitionMesh(): sorting [base nodes | higher order nodes] took 5.21e-07 s
[0] info: partitionMesh(): setting number of nodes and of all mesh base nodes took 1e-07 s
[0] info: partitionMesh(): distribute elements into partitions took 3e-07 s
[0] info: partitionMesh(): determine / append ghost nodes took 3.51e-07 s
[0] info: partitionMesh(): markDuplicateGhostCells took 1.8e-07 s
[0] info: Writing the partitions data into binary files took 0.00138338 s
[0] info: Total runtime: 0.11539 s.
[0] info: Total CPU time: 0.066698 s.
mesh path: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/mesh_GreatCell_VPF
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/ogs/Tests/Data/HMPhaseField/GreatCell
[DEBUG] output project file: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2a_VPF_A.prj
============================================================
Running simulation for load case: A with method: VPF, tension_cutoff: False
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Gneiss' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2a_VPF_A.prj
Status: finished successfully.
Execution took 85.966637134552 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
DEBUG: Setting ls to 0.002 based on mesh size 0.001
ls 0.002
Material properties for 'Greywacke' (VPF) applied successfully.
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/F1XUyv4cx/0/ogs/build/release-petsc/Tests/Data/HMPhaseField/GreatCell/GreatCellHM_VPF/HM2a_VPF_A.prj
Status: finished successfully.
Execution took 99.3064694404602 s
Post-processing
plotter = Plotter(…
(click to toggle)
plotter = Plotter(
output_dir=out_dir,
save_extracted_data=True,
)
data_dir = Path("external_data")
external_data = Plotter.load_external_data(data_dir, benchmark_tag="HM2a")
plotter.plot_volumetric_strain_vs_angle(
vtu_files_dict_full_HM,
model_type="HM2a",
ylim_range=[-7.5, 2.5],
layout="subplots",
external_data=external_data["strain"],
)[0] info: Reading meshes took 0.014461889000000117 s
[0] info: Collection of 2921 regular elements and computing element map took 1.92689999998219e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.1350000000429645e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0012190530000000255 s
[0] info: Reset nodes in regular elements took 0.005192171000000023 s
[0] info: creation of merged mesh took 0.00047827300000014006 s
[0] info: merge properties into merged mesh took 0.0003454019999997726 s
[0] info: writing mesh took 0.01447516800000015 s
[0] info: Reading meshes took 0.012316605000000092 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9419000000020503e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.084999999902749e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.001127395000000142 s
[0] info: Reset nodes in regular elements took 0.005342237999999888 s
[0] info: creation of merged mesh took 0.0004624489999998094 s
[0] info: merge properties into merged mesh took 0.0003534139999998853 s
[0] info: writing mesh took 0.013275574999999984 s

plotter.plot_field_variables(vtu_files_dict_full_HM)Load: A, File 0, Benchmark: HM2a, Material: Gneiss
[0] info: Reading meshes took 0.013519123000000022 s
[0] info: Collection of 2921 regular elements and computing element map took 1.9809999999953476e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.0240000001846568e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0011229480000001235 s
[0] info: Reset nodes in regular elements took 0.005308727000000069 s
[0] info: creation of merged mesh took 0.00047518899999987596 s
[0] info: merge properties into merged mesh took 0.0003532939999999485 s
[0] info: writing mesh took 0.012431148000000114 s

Load: A, File 1, Benchmark: HM2a, Material: Greywacke
[0] info: Reading meshes took 0.013624973999999845 s
[0] info: Collection of 2921 regular elements and computing element map took 2.060199999998069e-05 s
[0] info: Collection of 2879 nodes and computing offsets took 3.0450000001458477e-06 s
[0] info: Make nodes unique (2785 unique nodes) / computing map took 0.0012458940000001917 s
[0] info: Reset nodes in regular elements took 0.0052247599999999395 s
[0] info: creation of merged mesh took 0.0004760100000000822 s
[0] info: merge properties into merged mesh took 0.0003506710000000801 s
[0] info: writing mesh took 0.013137986999999907 s

plotter.material_names = ["Gneiss", "Greywacke"]…
(click to toggle)
plotter.material_names = ["Gneiss", "Greywacke"]
plotter.vtu_file_names = {"VPF": vtu_files_dict_full_HM}
plotter.plot_avg_width_vs_stress(
benchmark_tag="HM2a",
metric="width",
methods_to_include=["VPF"],
pee_load_values=PEE_load_values,
external_data=external_data["average"],
ylim_range=(0, 5.0e-5),
)
plotter.plot_avg_width_vs_stress(
benchmark_tag="HM2a",
metric="permeability",
methods_to_include=["VPF"],
pee_load_values=PEE_load_values,
external_data=external_data["average"],
ylim_range=(0, 2.5e-10),
)


array([1000000.])
vpf_profiles = {}…
(click to toggle)
vpf_profiles = {}
for load_case, file_list in plotter.vtu_file_names["VPF"].items():
vpf_profiles[load_case] = plotter.extract_vpf_width_from_list(
file_list,
plotter.material_names,
)
plotter.plot_fracture_aperture_profiles(
widthProfile=vpf_profiles,
benchmark_tag="HM2a",
downsample=1,
ylim=(0, 5.0e-5),
method_label="VPF",
external_data=external_data["widthProfile"],
)

pairs_to_check = {…
(click to toggle)
pairs_to_check = {
"M1_VPF_A_Greywacke": "M1",
"M2a_VPF_A_Greywacke": "M2a",
"M2b_VPF_A_Greywacke": "M2b",
}
for case, label in pairs_to_check.items():
print(f"\n===== {label} case =====")
new_result = np.load(Path(out_dir, f"extracted_{case}_volStrain.npz"))
expected_result = np.load(Path("expected", f"extracted_{case}_volStrain.npz"))
eps_v_new = new_result["eps_v"]
eps_v_expected = expected_result["eps_v"]
phi_new = new_result["phi"]
phi_expected = expected_result["phi"]
print(
f"eps_v_new shape: {eps_v_new.shape}, eps_v_expected shape: {eps_v_expected.shape}"
)
print(f"phi_new shape: {phi_new.shape}, phi_expected shape: {phi_expected.shape}")
np.testing.assert_allclose(eps_v_new, eps_v_expected, atol=5e-4)
np.testing.assert_allclose(phi_new, phi_expected, atol=1e-8)
print(f"\n{label} case passed.")===== M1 case =====
eps_v_new shape: (88,), eps_v_expected shape: (88,)
phi_new shape: (88,), phi_expected shape: (88,)
M1 case passed.
===== M2a case =====
eps_v_new shape: (74,), eps_v_expected shape: (74,)
phi_new shape: (74,), phi_expected shape: (74,)
M2a case passed.
===== M2b case =====
eps_v_new shape: (74,), eps_v_expected shape: (74,)
phi_new shape: (74,), phi_expected shape: (74,)
M2b case passed.
Reference
-
McDermott, C.I., Fraser-Harris, A., Sauter, M., Couples, G.D., Edlmann, K., Kolditz, O., Lightbody, A., Somerville, J. and Wang, W., 2018. New experimental equipment recreating geo-reservoir conditions in large, fractured, porous samples to investigate coupled thermal, hydraulic and polyaxial stress processes. Scientific reports, 8(1), p.14549.
-
Mollaali, M., Kolditz, O., Hu, M., Park, C.H., Park, J.W., McDermott, C.I., Chittenden, N., Bond, A., Yoon, J.S., Zhou, J. and Pan, P.Z., Liu H., Hou W., Lei H., Zhang L., Nagel T., Barsch M., Wang W., Nguyen S., Kwon S. and Yoshioka K., 2023. Comparative verification of hydro-mechanical fracture behavior: Task G of international research project DECOVALEX–2023. International Journal of Rock Mechanics and Mining Sciences, 170, p.105530.
-
Mollaali, M., Wang, W., You, T., Nagel, T., Fraser-Harris, A., McDermott, C., and Kolditz, O., 2025. Numerical benchmarking of GREAT cell experiments: Poly-axial stress effects on fluid flow in fractured rock using smeared and discrete methods.
This article was written by Mostafa Mollaali. If you are missing something or you find an error please let us know.
Generated with Hugo 0.147.9
in CI job 635984
|
Last revision: April 3, 2025