A 2D GREAT cell benchmark suite simulated using small deformation and LIE mechanical processes
import os…
(click to toggle)
import os
from pathlib import Path
from subprocess import run
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import ogstools as ot
from external_data_dict import external_data
from matplotlib import colormaps
# Local modules
from mesh_generator import (
mesh_GreatCell_embeddedFracture,
mesh_GreatCell_fullFracture,
mesh_GreatCell_intact,
)
from ogs_model_runner import SingleOGSModel
from Plotter import Plotter
def truncated_cmap(name, minval=0.3, maxval=0.9, n=6):…
(click to toggle)
def truncated_cmap(name, minval=0.3, maxval=0.9, n=6):
base = colormaps.get_cmap(name)
return lambda i: base(minval + (maxval - minval) * i / (n - 1))
mpl.rcdefaults()
mpl.rcParams.update(
{
"text.usetex": False,
"font.family": "serif",
"font.serif": ["DejaVu Serif"],
"axes.labelsize": 26,
"axes.titlesize": 24,
"legend.fontsize": 18,
"xtick.labelsize": 18,
"ytick.labelsize": 18,
}
)
ot.plot.setup.show_region_bounds = False…
(click to toggle)
ot.plot.setup.show_region_bounds = False
out_dir = Path(os.environ.get("OGS_TESTRUNNER_OUT_DIR", "_out"))
out_dir.mkdir(parents=True, exist_ok=True)Great cell

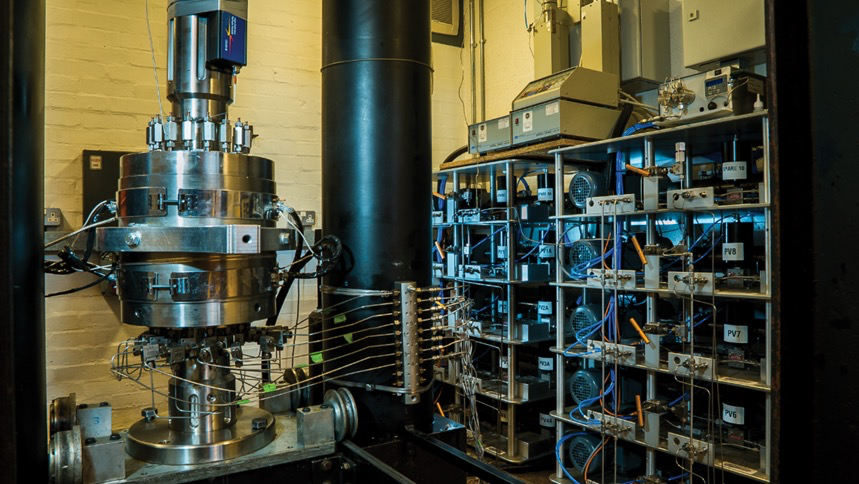
Figure: The GREAT cell facility.
The GREAT cell is designed to reproduce subsurface conditions in the laboratory down to a depth of 3.5 km, simulating both geomechanical stress and geothermal reservoir conditions on 200 mm diameter rock samples that contain fracture networks. This enables validation of numerical predictions under realistic conditions.
The GREAT cell represents a major advancement in experimental geomechanics technology. It uniquely:
- Generates a truly poly-axial rotating stress field
- Facilitates fluid flow through the rock samples
- Employs state-of-the-art fibre-optic strain sensing for high-resolution circumferential strain measurements
- Records fluid pressure and strain at thousands of data points per hour
This facility enhances understanding of fracture propagation and its effect on fluid flow—key factors in geo-energy applications involving fluid storage or extraction in the subsurface.

(a) Typical rock sample

(b) Simplification for GREAT cell benchmarking
Figure: (a) Typical rock sample and (b) its simplified 3D schematic used for GREAT cell benchmarks. The cross-section (b) aligns with the 2D simulations discussed earlier.
Benchmarking Strategy
A comprehensive benchmarking exercise is performed to establish the models’ capabilities and required settings, such as the required discretization for comparable accuracy. The exercise benchmarks are simplified versions of the GREAT cell experiments, focusing on general features like fracture patterns, rotating boundary conditions, and coupled processes.
These benchmark exercises are conducted in 2D (plane strain), representing a horizontal section through the middle of the rock samples tested in the GREAT cell (see Figure 1). This 2D plane-strain approach allows the main characteristics of hydro-mechanical (HM) fracture mechanics to be studied efficiently. The benchmarking concept involves a stepwise increase in complexity, including both mechanical (M) and hydro-mechanical (HM) approaches.
We distinguish two main types of benchmarks: intact and fractured rock tests (see Figure 2). In the first type, an intact rock sample is subjected to rotating external stress conditions ($\texttt{M}_1$) and fluid is injected from a central borehole ($\texttt{HM}_1$):
- $\texttt{M}_1$: Intact rock sample under rotating boundary conditions.
- $\texttt{HM}_1$: Intact rock sample with fluid injection.
For the fractured rock samples, two subtypes are also considered: static and propagating fractures. Static fractures are studied as full ($\texttt{M}_{2a}$, $\texttt{HM}_{2a}$) and half-fractured ($\texttt{M}_{2b}$, $\texttt{HM}_{2b}$) versions to have a symmetric and non-symmetric case. Again, mechanical and hydro-mechanical versions are studied, the latter with fluid injection into the rock fracture:
- $\texttt{M}_{2a}$: Fully fractured sample.
- $\texttt{M}_{2b}$: Half-fractured sample.
- $\texttt{HM}_{2a}$: Fully fractured sample with inflow-outflow.
- $\texttt{HM}_{2b}$: Half-fractured sample with inflow-outflow.
Finally, fracture propagation conditions are studied by injecting fluid into a pre-existing fracture ($\texttt{HM}_{3c}$) and by initiating a fracture from a borehole ($\texttt{HM}_{3d}$):
- $\texttt{HM}_{3c}$: Propagation of pre-existing fracture by fluid injection.
- $\texttt{HM}_{3d}$: Fracture nucleation from borehole by fluid injection.

Figure 1:
Three sample types of GREAT cell benchmarks: intact, fractured (static), and propagating. Rotating boundary conditions are applied.

Figure 2:
Geometry and location of PEEs and DSSs. Fracture aligned with PEE1/1a direction.
Material Properties
The material properties are provided in Table 1. The computational model incorporates two distinct elastic materials within its domain: a central circle ($r=0.097$ m) of rock surrounded by a rubber sheath in a 2D configuration.
Table 1: Material Properties
| Parameter | Unit | Rubber Sheath | Greywacke | Gneiss (Freiberg) |
|---|---|---|---|---|
| Young’s modulus, $E$ | GPa | 0.1 | 26.85 | 83.9 |
| Poisson’s ratio, $\nu$ | - | 0.4 | 0.27 | 0.21 |
| Tensile strength | MPa | - | 17.01–16.67 | 16.8 |
| Permeability, $K_m$ | m² | $1 \times 10^{-19}$ | $2.58 \times 10^{-19}$ | $1 \times 10^{-19}$ |
| Solid density, $\rho_s$ | kg/m³ | 1200 | 2650 | 2750 |
| Porosity, $\phi$ | - | - | 0.005 | 0.001 |
| Biot coefficient, $\alpha_m$ | - | 0.0 | 0.8 | 0.6 |
| Fracture energy, $G_c$ (VPF-FEM) | N/m | 0.1 | 30 | 50 |
| Normal stiffness, $k_{nn}$ (LIE-FEM) | GPa/m | - | 100.0 | 200.0 |
| Tangential stiffness, $k_{tt}$ (LIE-FEM) | GPa/m | - | 50.0 | 100.0 |
materials = {…
(click to toggle)
materials = {
"Gneiss": {
"young_sample": 83.9e9, # Young's modulus (Pa)
"nu_sample": 0.21, # Poisson's ratio
"biot": 0.6, # Biot coefficient
"porosity": 0.001, # Porosity
"permeability": 1e-19, # Permeability (m²)
"density_solid": 2750, # Solid density (kg/m³)
"k_n": 200e9, # Normal stiffness (Pa/m)
"k_t": 100e9, # Tangential stiffness (Pa/m)
"c_f": 4.4e-10, # Fluid compressibility (Pa⁻¹)
"k_s": 4.82e10, # Solid bulk modulus (Pa)
"S_f": 4.4e-10, # Specific storage (Pa⁻¹)
"t_np": 10e6, # Peak normal traction (Pa)
"Gc": 50, # Fracture toughness (J/m²)
"w_init": 1e-6, # initial fracture width (m)
"rubber_sheath": {
"young_modulus": 0.1e9, # Young's modulus (Pa)
"poisson_ratio": 0.4, # Poisson's ratio
"porosity": 0.001, # Porosity
"permeability": 1e-17, # Permeability (m²)
"density": 1500, # Density (kg/m³)
"biot": 0.0, # Biot coefficient
},
},
"Greywacke": {
"young_sample": 26.87e9, # Young's modulus (Pa)
"nu_sample": 0.27, # Poisson's ratio
"biot": 0.8, # Biot coefficient
"porosity": 0.005, # Porosity
"permeability": 2.58e-19, # Permeability (m²)
"density_solid": 2650, # Solid density (kg/m³)
"k_n": 100e9, # Normal stiffness (Pa/m)
"k_t": 50e9, # Tangential stiffness (Pa/m)
"c_f": 4.4e-10, # Fluid compressibility (Pa⁻¹)
"k_s": 1.95e10, # Solid bulk modulus (Pa)
"S_f": 4.4e-10, # Specific storage (Pa⁻¹)
"t_np": 10e6, # Peak normal traction (Pa)
"Gc": 30, # Fracture toughness (J/m²)
"w_init": 1e-6, # initial fracture width (m)
"rubber_sheath": {
"young_modulus": 0.1e9, # Young's modulus (Pa)
"poisson_ratio": 0.4, # Poisson's ratio
"porosity": 0.001, # Porosity
"permeability": 1e-17, # Permeability (m²)
"density": 1500, # Density (kg/m³)
"biot": 0.0, # Biot coefficient
},
},
}
material_names = list(materials.keys())Loads
Table 2: Load Conditions
| Marker | PEE1 Angle to $\sigma_2$ | PEE1 & 1a | PEE2 & 2a | PEE3 & 3a | PEE4 & 4a | PEE5 & 5a | PEE6 & 6a | PEE7 & 7a | PEE8 & 8a |
|---|---|---|---|---|---|---|---|---|---|
| A | 0° | 10.0 | 6.64 | 4.46 | 1.17 | 1.0 | 3.82 | 7.80 | 9.95 |
| E | 22.5° | 9.95 | 10.0 | 6.64 | 4.46 | 1.17 | 1.0 | 3.82 | 7.80 |
| B | 45.0° | 7.80 | 9.95 | 10.0 | 6.64 | 4.46 | 1.17 | 1.0 | 3.82 |
| F | 67.5° | 3.82 | 7.80 | 9.95 | 10.0 | 6.64 | 4.46 | 1.17 | 1.0 |
| C | 90° | 1.0 | 3.82 | 7.80 | 9.95 | 10.0 | 6.64 | 4.46 | 1.17 |
All loads are in MPa. DSS loads are averages of adjacent PEEs.
Schematic loading conditions
loads = {…
(click to toggle)
loads = {
"A": [
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
7.80e6,
9.95e6,
],
"B": [
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
],
"C": [
1.0e6,
3.82e6,
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
],
}
angles_degrees = […
(click to toggle)
angles_degrees = [
0,
-22.5,
-45,
-67.5,
-90,
-112.5,
-135,
-157.5,
-180,
-202.5,
-225,
-247.5,
-270,
-292.5,
-315,
-337.5,
]
angles_radians = np.deg2rad(angles_degrees)
circle_radius = 6
circle_x = circle_radius * np.cos(angles_radians)
circle_y = circle_radius * np.sin(angles_radians)
fig, axs = plt.subplots(1, 3, figsize=(21, 7), facecolor="none")
for idx, (label, values) in enumerate(loads.items()):
ax = axs[idx]
ax.set_aspect("equal")
ax.axis("off")
formatted_values = [rf"${v / 1e6:.1f}$" for v in values]
scaled_values = [v / 2 for v in values]
circle = plt.Circle(
(0, 0),
circle_radius,
color="black",
fill=False,
linestyle="--",
linewidth=2,
)
ax.add_artist(circle)
top_points_x, top_points_y = [], []
for i, (x, y, value) in enumerate(
zip(circle_x, circle_y, scaled_values, strict=True)
):
unit_vector = np.array([x, y]) / circle_radius
line_end = np.array([x, y]) + unit_vector * value / 1e6
top_points_x.append(line_end[0])
top_points_y.append(line_end[1])
ax.annotate(
"",
xytext=line_end,
xy=(x, y),
arrowprops={
"arrowstyle": "-|>",
"color": "blue",
"lw": 2,
"mutation_scale": 15,
"fill": True,
},
)
angle_offset = 1.2 * unit_vector
angle_label = f"{-angles_degrees[i]}°"
ax.text(
x - angle_offset[0],
y - angle_offset[1],
angle_label,
fontsize=12,
ha="center",
va="center",
color="m",
)
value_offset = 0.5 * unit_vector
ax.text(
line_end[0] + 3.0 * value_offset[0],
line_end[1] + 1.8 * value_offset[1],
formatted_values[i],
fontsize=18,
ha="center",
weight="bold",
)
top_points_x.append(top_points_x[0])
top_points_y.append(top_points_y[0])
ax.plot(
top_points_x,
top_points_y,
color="green",
linestyle="-.",
lw=2,
marker="o",
markersize=6,
markerfacecolor="lightgreen",
markeredgewidth=1.0,
markeredgecolor="black",
)
ax.set_xlim([-12, 12])
ax.set_ylim([-12, 12])
ax.text(
0,
0,
f"Load {label}",
fontsize=32,
ha="center",
va="center",
family="serif",
)
plt.tight_layout()
output_path = Path(out_dir, "loads_A_B_C_schematic_with_angles.png")
plt.savefig(output_path, dpi=350, bbox_inches="tight", transparent=True)
plt.show()

Boundary Conditions
The boundary conditions applied in the simulation include both Dirichlet and Neumann conditions.
- Dirichlet conditions (displacement control):
\begin{equation*} \begin{cases} u_x(0, y) = 0, \quad u_y(0, y) = 0 & \quad \text{for } y = -0.09894 \text{ m}, \ u_y(x, 0) = 0, & \quad \text{for } x = -0.09894 \text{ m}. \end{cases} \end{equation*}
- Neumann conditions: Normal stress $\sigma_{rr}$ is applied on PEEs and DSSs. The DSS stress is calculated as: $$\sigma_\text{DSS}^i = \frac{\sigma_\text{PEE}^i + \sigma_\text{PEE}^{i+1}}{2}$$
Intact Samples ($\texttt{M}_1$)
The primary objective of the first benchmark exercise is to evaluate the mechanical deformation of different rock samples in the GREAT cell under various boundary conditions in a 2D plane strain setup.
This benchmark considers a plane strain triaxial stress condition, where the in-plane principal stresses are defined as:
$$ \sigma_2 = 10\ \text{MPa},\quad \sigma_3 = 1\ \text{MPa} $$The out-of-plane stress component, $\sigma_1$, is governed by the plane strain constraint:
$$ \sigma_1 = \nu (\sigma_2 + \sigma_3) $$This loading condition is applied to both Greywacke and Gneiss samples (see also loading Table referenced in earlier section).
Mesh Generation
Input
h = 0.0025…
(click to toggle)
h = 0.0025
meshname = "GreatCell"
mesh_path = Path(out_dir, "mesh_intact").resolve()
print(mesh_path)/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact
Boundary meshes
Gmsh (boundary meshes)
msh_file = mesh_GreatCell_intact(…
(click to toggle)
msh_file = mesh_GreatCell_intact(
lc=0.005,
lc2=h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path,
meshname=meshname,
mode="BC",
post_process=True,
cmap="viridis",
opacity=0.9,
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 20%] Meshing curve 110 (Line)
Info : [ 20%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 30%] Meshing curve 118 (Line)
Info : [ 30%] Meshing curve 119 (Line)
Info : [ 30%] Meshing curve 120 (Line)
Info : [ 30%] Meshing curve 121 (Line)
Info : [ 30%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 40%] Meshing curve 126 (Line)
Info : [ 40%] Meshing curve 127 (Line)
Info : [ 40%] Meshing curve 128 (Line)
Info : [ 40%] Meshing curve 129 (Line)
Info : [ 40%] Meshing curve 130 (Line)
Info : [ 40%] Meshing curve 131 (Line)
Info : [ 40%] Meshing curve 132 (Line)
Info : [ 40%] Meshing curve 133 (Line)
Info : [ 50%] Meshing curve 134 (Line)
Info : [ 50%] Meshing curve 135 (Line)
Info : [ 50%] Meshing curve 136 (Line)
Info : [ 50%] Meshing curve 137 (Line)
Info : [ 50%] Meshing curve 138 (Line)
Info : [ 50%] Meshing curve 139 (Line)
Info : [ 50%] Meshing curve 140 (Line)
Info : [ 50%] Meshing curve 141 (Line)
Info : [ 60%] Meshing curve 142 (Line)
Info : [ 60%] Meshing curve 143 (Line)
Info : [ 60%] Meshing curve 144 (Line)
Info : [ 60%] Meshing curve 145 (Line)
Info : [ 60%] Meshing curve 146 (Line)
Info : [ 60%] Meshing curve 147 (Line)
Info : [ 60%] Meshing curve 148 (Line)
Info : [ 60%] Meshing curve 149 (Line)
Info : [ 70%] Meshing curve 150 (Line)
Info : [ 70%] Meshing curve 151 (Line)
Info : [ 70%] Meshing curve 152 (Line)
Info : [ 70%] Meshing curve 153 (Line)
Info : [ 70%] Meshing curve 154 (Line)
Info : [ 70%] Meshing curve 155 (Line)
Info : [ 70%] Meshing curve 156 (Line)
Info : [ 70%] Meshing curve 157 (Line)
Info : [ 80%] Meshing curve 158 (Line)
Info : [ 80%] Meshing curve 159 (Line)
Info : [ 80%] Meshing curve 160 (Line)
Info : [ 80%] Meshing curve 161 (Line)
Info : [ 80%] Meshing curve 162 (Line)
Info : [ 80%] Meshing curve 163 (Line)
Info : [ 80%] Meshing curve 164 (Line)
Info : [ 80%] Meshing curve 1234 (Line)
Info : [ 90%] Meshing curve 3005 (Circle)
Info : [ 90%] Meshing curve 3006 (Circle)
Info : [ 90%] Meshing curve 3007 (Circle)
Info : [ 90%] Meshing curve 3008 (Circle)
Info : [ 90%] Meshing curve 3015 (Circle)
Info : [ 90%] Meshing curve 3016 (Circle)
Info : [ 90%] Meshing curve 3017 (Circle)
Info : [ 90%] Meshing curve 3018 (Circle)
Info : [100%] Meshing curve 3025 (Circle)
Info : [100%] Meshing curve 3026 (Circle)
Info : [100%] Meshing curve 3027 (Circle)
Info : [100%] Meshing curve 3028 (Circle)
Info : [100%] Meshing curve 3035 (Circle)
Info : [100%] Meshing curve 3036 (Circle)
Info : [100%] Meshing curve 3037 (Circle)
Info : [100%] Meshing curve 3038 (Circle)
Info : Done meshing 1D (Wall 0.00473061s, CPU 0.004695s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 200 (Plane, Delaunay)
Info : [ 0%] Blossom: 8705 internal 168 closed
Info : [ 0%] Blossom recombination completed (Wall 0.160914s, CPU 0.081111s): 2854 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.769982, min Q = 0.452527
Info : [ 30%] Meshing surface 201 (Plane, Delaunay)
Info : [ 30%] Blossom: 8008 internal 400 closed
Info : [ 30%] Blossom recombination completed (Wall 0.0951049s, CPU 0.06481s): 2590 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.791636, min Q = 0.499656
Info : [ 50%] Meshing surface 202 (Plane, Delaunay)
Info : [ 50%] Blossom: 1168 internal 472 closed
Info : [ 50%] Blossom recombination completed (Wall 0.00915129s, CPU 0.006125s): 468 quads, 0 triangles, 0 invalid quads, 4 quads with Q < 0.1, avg Q = 0.749983, min Q = 0.0166667
Info : [ 70%] Meshing surface 203 (Plane, Delaunay)
Info : [ 70%] Blossom: 1208 internal 488 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00864798s, CPU 0.009137s): 480 quads, 8 triangles, 0 invalid quads, 4 quads with Q < 0.1, avg Q = 0.672659, min Q = 0.016129
Info : [ 90%] Meshing surface 204 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.458645s, CPU 0.276467s)
Info : 6777 nodes 8146 elements
Info : Writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/GreatCell.msh'
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/domain.vtu (160 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_Inlet.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_p_right.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_p_top.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_p_left.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_p_bottom.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE1.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE2.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE3.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE4.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE5.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE6.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE7.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE8.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE1a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE2a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE3a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE4a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE5a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE6a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE7a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_PEE8a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS1.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS2.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS3.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS4.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS5.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS6.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS7.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS8.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS1a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS2a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS3a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS4a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS5a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS6a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS7a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_DSS8a.vtu (4 cells)
[0m[33m2025-10-23 14:27:24.023 ( 3.284s) [ 7FBD07C4C400]vtkXOpenGLRenderWindow.:1416 WARN| bad X server connection. DISPLAY=[0m

Computational domain mesh
Gmsh
msh_file = mesh_GreatCell_intact(…
(click to toggle)
msh_file = mesh_GreatCell_intact(
lc=0.005,
lc2=h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path,
meshname=meshname,
mode="domain",
post_process=True,
cmap="viridis",
opacity=0.8,
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 20%] Meshing curve 110 (Line)
Info : [ 20%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 30%] Meshing curve 118 (Line)
Info : [ 30%] Meshing curve 119 (Line)
Info : [ 30%] Meshing curve 120 (Line)
Info : [ 30%] Meshing curve 121 (Line)
Info : [ 30%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 40%] Meshing curve 126 (Line)
Info : [ 40%] Meshing curve 127 (Line)
Info : [ 40%] Meshing curve 128 (Line)
Info : [ 40%] Meshing curve 129 (Line)
Info : [ 40%] Meshing curve 130 (Line)
Info : [ 40%] Meshing curve 131 (Line)
Info : [ 40%] Meshing curve 132 (Line)
Info : [ 40%] Meshing curve 133 (Line)
Info : [ 50%] Meshing curve 134 (Line)
Info : [ 50%] Meshing curve 135 (Line)
Info : [ 50%] Meshing curve 136 (Line)
Info : [ 50%] Meshing curve 137 (Line)
Info : [ 50%] Meshing curve 138 (Line)
Info : [ 50%] Meshing curve 139 (Line)
Info : [ 50%] Meshing curve 140 (Line)
Info : [ 50%] Meshing curve 141 (Line)
Info : [ 60%] Meshing curve 142 (Line)
Info : [ 60%] Meshing curve 143 (Line)
Info : [ 60%] Meshing curve 144 (Line)
Info : [ 60%] Meshing curve 145 (Line)
Info : [ 60%] Meshing curve 146 (Line)
Info : [ 60%] Meshing curve 147 (Line)
Info : [ 60%] Meshing curve 148 (Line)
Info : [ 60%] Meshing curve 149 (Line)
Info : [ 70%] Meshing curve 150 (Line)
Info : [ 70%] Meshing curve 151 (Line)
Info : [ 70%] Meshing curve 152 (Line)
Info : [ 70%] Meshing curve 153 (Line)
Info : [ 70%] Meshing curve 154 (Line)
Info : [ 70%] Meshing curve 155 (Line)
Info : [ 70%] Meshing curve 156 (Line)
Info : [ 70%] Meshing curve 157 (Line)
Info : [ 80%] Meshing curve 158 (Line)
Info : [ 80%] Meshing curve 159 (Line)
Info : [ 80%] Meshing curve 160 (Line)
Info : [ 80%] Meshing curve 161 (Line)
Info : [ 80%] Meshing curve 162 (Line)
Info : [ 80%] Meshing curve 163 (Line)
Info : [ 80%] Meshing curve 164 (Line)
Info : [ 80%] Meshing curve 1234 (Line)
Info : [ 90%] Meshing curve 3005 (Circle)
Info : [ 90%] Meshing curve 3006 (Circle)
Info : [ 90%] Meshing curve 3007 (Circle)
Info : [ 90%] Meshing curve 3008 (Circle)
Info : [ 90%] Meshing curve 3015 (Circle)
Info : [ 90%] Meshing curve 3016 (Circle)
Info : [ 90%] Meshing curve 3017 (Circle)
Info : [ 90%] Meshing curve 3018 (Circle)
Info : [100%] Meshing curve 3025 (Circle)
Info : [100%] Meshing curve 3026 (Circle)
Info : [100%] Meshing curve 3027 (Circle)
Info : [100%] Meshing curve 3028 (Circle)
Info : [100%] Meshing curve 3035 (Circle)
Info : [100%] Meshing curve 3036 (Circle)
Info : [100%] Meshing curve 3037 (Circle)
Info : [100%] Meshing curve 3038 (Circle)
Info : Done meshing 1D (Wall 0.00549967s, CPU 0.004546s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 200 (Plane, Delaunay)
Info : [ 0%] Blossom: 8705 internal 168 closed
Info : [ 0%] Blossom recombination completed (Wall 0.0899661s, CPU 0.074434s): 2854 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.769982, min Q = 0.452527
Info : [ 30%] Meshing surface 201 (Plane, Delaunay)
Info : [ 30%] Blossom: 8008 internal 400 closed
Info : [ 30%] Blossom recombination completed (Wall 0.0750779s, CPU 0.059607s): 2590 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.791636, min Q = 0.499656
Info : [ 50%] Meshing surface 202 (Plane, Delaunay)
Info : [ 50%] Blossom: 1168 internal 472 closed
Info : [ 50%] Blossom recombination completed (Wall 0.0140435s, CPU 0.005506s): 468 quads, 0 triangles, 0 invalid quads, 4 quads with Q < 0.1, avg Q = 0.749983, min Q = 0.0166667
Info : [ 70%] Meshing surface 203 (Plane, Delaunay)
Info : [ 70%] Blossom: 1208 internal 488 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00842478s, CPU 0.008391s): 480 quads, 8 triangles, 0 invalid quads, 4 quads with Q < 0.1, avg Q = 0.672659, min Q = 0.016129
Info : [ 90%] Meshing surface 204 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.313479s, CPU 0.256214s)
Info : 6777 nodes 8146 elements
Info : Writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/GreatCell.msh'
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/domain.vtu (7000 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_Central_sample.vtu (2854 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_OuterPart_sample.vtu (3546 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact/physical_group_Rubber_sheath.vtu (600 cells)

%cd {mesh_path}…
(click to toggle)
%cd {mesh_path}
run(
"identifySubdomains -f -m domain.vtu -- "
"physical_group_DSS1.vtu physical_group_DSS1a.vtu "
"physical_group_DSS2.vtu physical_group_DSS2a.vtu "
"physical_group_DSS3.vtu physical_group_DSS3a.vtu "
"physical_group_DSS4.vtu physical_group_DSS4a.vtu "
"physical_group_DSS5.vtu physical_group_DSS5a.vtu "
"physical_group_DSS6.vtu physical_group_DSS6a.vtu "
"physical_group_DSS7.vtu physical_group_DSS7a.vtu "
"physical_group_DSS8.vtu physical_group_DSS8a.vtu "
"physical_group_PEE1.vtu physical_group_PEE1a.vtu "
"physical_group_PEE2.vtu physical_group_PEE2a.vtu "
"physical_group_PEE3.vtu physical_group_PEE3a.vtu "
"physical_group_PEE4.vtu physical_group_PEE4a.vtu "
"physical_group_PEE5.vtu physical_group_PEE5a.vtu "
"physical_group_PEE6.vtu physical_group_PEE6a.vtu "
"physical_group_PEE7.vtu physical_group_PEE7a.vtu "
"physical_group_PEE8.vtu physical_group_PEE8a.vtu "
"physical_group_p_bottom.vtu physical_group_p_left.vtu "
"physical_group_p_right.vtu physical_group_p_top.vtu ",
shell=True,
check=True,
)
%cd -/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact
info: Mesh reading time: 0.00966455 s
info: MeshNodeSearcher construction time: 0.000298812 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.098e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000587077 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.31e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000270889 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.11e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000239372 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.61e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000233402 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.11e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000234064 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.22e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000236257 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.51e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000238901 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.31e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000233523 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.31e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000237038 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.91e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000232421 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.01e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000232041 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.41e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000235476 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.71e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000230288 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.31e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000229176 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.51e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000229296 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.51e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.00023149 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 2.103e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000237038 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.252e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000229927 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.162e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000217949 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000230809 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.082e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000236917 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.061e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000230628 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.12e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000230168 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.82e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000229867 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.52e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000296929 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.102e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000233413 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.101e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000235856 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.71e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000223768 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.72e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000237469 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.272e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000234334 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000237679 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.22e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000258681 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.3e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_bottom' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000234524 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.31e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_left' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000256248 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.1e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_right' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000237579 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 2.9e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_top' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.00023172 s
info: identifySubdomains time: 0.0103506 s
info: writing time: 0.0227994 s
info: Entire run time: 0.0435826 s
/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
Run the simulation
Inputs
# Times for load curves…
(click to toggle)
# Times for load curves
times = "0.0 1000. 3500"
simulation_end_time = 3500.0
n_fracture_p_ncs = 0
model_type = "M1"
output_prefix = "M1_SD"
# Load
PEE_load_values = {
"A": [10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6, 9.95e6],
"B": [7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6],
"C": [1.0e6, 3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6],
}
prj_file = Path("M1_SD.prj")
# Create a Project object
prj = ot.Project(input_file=prj_file, output_file=Path(out_dir, f"{output_prefix}.prj"))Run OGS
# Create SingleOGSModel…
(click to toggle)
# Create SingleOGSModel
sing_ogs_model = SingleOGSModel(
model=prj,
out_dir=out_dir,
mesh_path=mesh_path,
output_prefix=output_prefix,
method="LIE",
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
materials=materials,
)
# Run simulations
vtu_files_dict = sing_ogs_model.run_simulations_with_fracture(
times=times,
base_project_file=prj_file,
mesh_path=mesh_path,
load_cases=PEE_load_values,
material_names=material_names,
materials=materials,
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
output_prefix=output_prefix,
out_dir=out_dir,
use_b_bar_value="true",
tension_cutoff="true",
)mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M1_SD.prj
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M1_SD_A.prj
============================================================
Running simulation for load case: A with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M1_SD_A.prj
Status: finished successfully.
Execution took 26.880229473114014 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M1_SD_A.prj
Status: finished successfully.
Execution took 32.9832124710083 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M1_SD_B.prj
============================================================
Running simulation for load case: B with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M1_SD_B.prj
Status: finished successfully.
Execution took 22.151084423065186 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M1_SD_B.prj
Status: finished successfully.
Execution took 28.47843623161316 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M1_SD_C.prj
============================================================
Running simulation for load case: C with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M1_SD_C.prj
Status: finished successfully.
Execution took 16.796292781829834 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M1_SD_C.prj
Status: finished successfully.
Execution took 19.897427797317505 s
Post-processing
Volumetric strain vs angle at probe circle
json_path = Path("./external_data_dict.py").resolve()…
(click to toggle)
json_path = Path("./external_data_dict.py").resolve()
print(f"[DEBUG] Trying path: {json_path}")
if json_path.exists():
from external_data_dict import external_data
else:
print("[WARNING] External data dict not found! Skipping...")
external_data = None
plotter = Plotter(
output_dir=out_dir,
save_extracted_data=True,
)
plotter.plot_volumetric_strain_vs_angle(
vtu_files_dict,
model_type="M1",
ylim_range=[-7.5, 2.5],
layout="subplots",
external_data=external_data,
)[DEBUG] Trying path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/external_data_dict.py

Profiles
Figure below shows the results of this benchmark:
- The trace of effective stress, defined as $\sum_{i=x,y,z} \sigma'_{ii}$,
- and the corresponding volumetric strain plotted versus loading angle,
- for both Greywacke and Gneiss,
These results are shown for the intact rock samples under $\texttt{M}_1$ loading.
plotter.plot_field_variables(vtu_files_dict)Load: A, File 0, Benchmark: M1, Material: Gneiss

Load: A, File 1, Benchmark: M1, Material: Greywacke

Load: B, File 0, Benchmark: M1, Material: Gneiss

Load: B, File 1, Benchmark: M1, Material: Greywacke

Load: C, File 0, Benchmark: M1, Material: Gneiss

Load: C, File 1, Benchmark: M1, Material: Greywacke

Note: The stress distribution and volumetric strain are shown only in the central zone defined by:
$$ \sqrt{x^2 + y^2} < 0.065\ \text{m} $$This is done because strain is relatively high in the PEE regions (due to their softer material properties), and we want to isolate the deformation behavior within the core of the sample.
# Plot only inner mesh (within r=0.065 m):
plotter.plot_field_variables(vtu_files_dict, inner=True, r=0.065)Load: A, File 0, Benchmark: M1, Material: Gneiss

Load: A, File 1, Benchmark: M1, Material: Greywacke

Load: B, File 0, Benchmark: M1, Material: Gneiss

Load: B, File 1, Benchmark: M1, Material: Greywacke

Load: C, File 0, Benchmark: M1, Material: Gneiss

Load: C, File 1, Benchmark: M1, Material: Greywacke

Half fractured sample ($\texttt{M}_{2b}$)
In this section, all input data and boundary conditions provided in $\texttt{M}_{2b}$, except a single wing fracture, defined as $\Gamma = \left[0, 0.04\right] \times \{0\}$, is considered.
Mesh Generation
Input
h = 0.0025…
(click to toggle)
h = 0.0025
meshname = "GreatCell"
mesh_path_embedded = Path(out_dir, "mesh_GreatCell_embeddedFracture").resolve()Boundary meshes
Gmsh (boundary meshes)
msh_file_embedded = mesh_GreatCell_embeddedFracture(…
(click to toggle)
msh_file_embedded = mesh_GreatCell_embeddedFracture(
lc=0.005,
lc2=h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path_embedded,
meshname=meshname,
mode="BC",
post_process=True,
cmap="viridis",
opacity=0.8,
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 20%] Meshing curve 110 (Line)
Info : [ 20%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 30%] Meshing curve 118 (Line)
Info : [ 30%] Meshing curve 119 (Line)
Info : [ 30%] Meshing curve 120 (Line)
Info : [ 30%] Meshing curve 121 (Line)
Info : [ 30%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 40%] Meshing curve 126 (Line)
Info : [ 40%] Meshing curve 127 (Line)
Info : [ 40%] Meshing curve 128 (Line)
Info : [ 40%] Meshing curve 129 (Line)
Info : [ 40%] Meshing curve 130 (Line)
Info : [ 40%] Meshing curve 131 (Line)
Info : [ 40%] Meshing curve 132 (Line)
Info : [ 40%] Meshing curve 133 (Line)
Info : [ 50%] Meshing curve 134 (Line)
Info : [ 50%] Meshing curve 135 (Line)
Info : [ 50%] Meshing curve 136 (Line)
Info : [ 50%] Meshing curve 137 (Line)
Info : [ 50%] Meshing curve 138 (Line)
Info : [ 50%] Meshing curve 139 (Line)
Info : [ 50%] Meshing curve 140 (Line)
Info : [ 50%] Meshing curve 141 (Line)
Info : [ 60%] Meshing curve 142 (Line)
Info : [ 60%] Meshing curve 143 (Line)
Info : [ 60%] Meshing curve 144 (Line)
Info : [ 60%] Meshing curve 145 (Line)
Info : [ 60%] Meshing curve 146 (Line)
Info : [ 60%] Meshing curve 147 (Line)
Info : [ 60%] Meshing curve 148 (Line)
Info : [ 60%] Meshing curve 149 (Line)
Info : [ 70%] Meshing curve 150 (Line)
Info : [ 70%] Meshing curve 151 (Line)
Info : [ 70%] Meshing curve 152 (Line)
Info : [ 70%] Meshing curve 153 (Line)
Info : [ 70%] Meshing curve 154 (Line)
Info : [ 70%] Meshing curve 155 (Line)
Info : [ 70%] Meshing curve 156 (Line)
Info : [ 70%] Meshing curve 157 (Line)
Info : [ 80%] Meshing curve 158 (Line)
Info : [ 80%] Meshing curve 159 (Line)
Info : [ 80%] Meshing curve 160 (Line)
Info : [ 80%] Meshing curve 161 (Line)
Info : [ 80%] Meshing curve 162 (Line)
Info : [ 80%] Meshing curve 163 (Line)
Info : [ 80%] Meshing curve 164 (Line)
Info : [ 80%] Meshing curve 1234 (Line)
Info : [ 90%] Meshing curve 3005 (Circle)
Info : [ 90%] Meshing curve 3006 (Circle)
Info : [ 90%] Meshing curve 3007 (Circle)
Info : [ 90%] Meshing curve 3008 (Circle)
Info : [ 90%] Meshing curve 3015 (Circle)
Info : [ 90%] Meshing curve 3016 (Circle)
Info : [ 90%] Meshing curve 3017 (Circle)
Info : [ 90%] Meshing curve 3018 (Circle)
Info : [100%] Meshing curve 3025 (Circle)
Info : [100%] Meshing curve 3026 (Circle)
Info : [100%] Meshing curve 3027 (Circle)
Info : [100%] Meshing curve 3028 (Circle)
Info : [100%] Meshing curve 3035 (Circle)
Info : [100%] Meshing curve 3036 (Circle)
Info : [100%] Meshing curve 3037 (Circle)
Info : [100%] Meshing curve 3038 (Circle)
Info : Done meshing 1D (Wall 0.00596452s, CPU 0.004451s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 200 (Plane, Delaunay)
Info : [ 0%] Blossom: 8705 internal 168 closed
Info : [ 0%] Blossom recombination completed (Wall 0.0749784s, CPU 0.074629s): 2854 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.769982, min Q = 0.452527
Info : [ 30%] Meshing surface 201 (Plane, Delaunay)
Info : [ 30%] Blossom: 8008 internal 400 closed
Info : [ 30%] Blossom recombination completed (Wall 0.062284s, CPU 0.061196s): 2590 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.791636, min Q = 0.499656
Info : [ 50%] Meshing surface 202 (Plane, Delaunay)
Info : [ 50%] Blossom: 1168 internal 472 closed
Info : [ 50%] Blossom recombination completed (Wall 0.00577876s, CPU 0.005741s): 468 quads, 0 triangles, 0 invalid quads, 4 quads with Q < 0.1, avg Q = 0.749983, min Q = 0.0166667
Info : [ 70%] Meshing surface 203 (Plane, Delaunay)
Info : [ 70%] Blossom: 1208 internal 488 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00869263s, CPU 0.008622s): 480 quads, 8 triangles, 0 invalid quads, 4 quads with Q < 0.1, avg Q = 0.672659, min Q = 0.016129
Info : [ 90%] Meshing surface 204 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.262416s, CPU 0.259267s)
Info : 6777 nodes 8146 elements
Info : Writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/GreatCell.msh'
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/domain.vtu (160 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_Inlet.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_Outlet_R.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_p_right.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_p_top.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_p_left.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_p_bottom.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE1.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE2.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE3.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE4.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE5.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE6.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE7.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE8.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE1a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE2a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE3a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE4a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE5a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE6a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE7a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_PEE8a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS1.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS2.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS3.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS4.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS5.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS6.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS7.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS8.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS1a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS2a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS3a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS4a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS5a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS6a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS7a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_DSS8a.vtu (4 cells)

Computational domain mesh
Gmsh (computational domain mesh)
msh_file_volume_embedded = mesh_GreatCell_embeddedFracture(…
(click to toggle)
msh_file_volume_embedded = mesh_GreatCell_embeddedFracture(
lc=0.005,
lc2=h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path_embedded,
meshname=meshname,
mode="domain",
post_process=True,
cmap="viridis",
opacity=0.8,
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 20%] Meshing curve 110 (Line)
Info : [ 20%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 30%] Meshing curve 118 (Line)
Info : [ 30%] Meshing curve 119 (Line)
Info : [ 30%] Meshing curve 120 (Line)
Info : [ 30%] Meshing curve 121 (Line)
Info : [ 30%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 40%] Meshing curve 126 (Line)
Info : [ 40%] Meshing curve 127 (Line)
Info : [ 40%] Meshing curve 128 (Line)
Info : [ 40%] Meshing curve 129 (Line)
Info : [ 40%] Meshing curve 130 (Line)
Info : [ 40%] Meshing curve 131 (Line)
Info : [ 40%] Meshing curve 132 (Line)
Info : [ 40%] Meshing curve 133 (Line)
Info : [ 50%] Meshing curve 134 (Line)
Info : [ 50%] Meshing curve 135 (Line)
Info : [ 50%] Meshing curve 136 (Line)
Info : [ 50%] Meshing curve 137 (Line)
Info : [ 50%] Meshing curve 138 (Line)
Info : [ 50%] Meshing curve 139 (Line)
Info : [ 50%] Meshing curve 140 (Line)
Info : [ 50%] Meshing curve 141 (Line)
Info : [ 60%] Meshing curve 142 (Line)
Info : [ 60%] Meshing curve 143 (Line)
Info : [ 60%] Meshing curve 144 (Line)
Info : [ 60%] Meshing curve 145 (Line)
Info : [ 60%] Meshing curve 146 (Line)
Info : [ 60%] Meshing curve 147 (Line)
Info : [ 60%] Meshing curve 148 (Line)
Info : [ 60%] Meshing curve 149 (Line)
Info : [ 70%] Meshing curve 150 (Line)
Info : [ 70%] Meshing curve 151 (Line)
Info : [ 70%] Meshing curve 152 (Line)
Info : [ 70%] Meshing curve 153 (Line)
Info : [ 70%] Meshing curve 154 (Line)
Info : [ 70%] Meshing curve 155 (Line)
Info : [ 70%] Meshing curve 156 (Line)
Info : [ 70%] Meshing curve 157 (Line)
Info : [ 80%] Meshing curve 158 (Line)
Info : [ 80%] Meshing curve 159 (Line)
Info : [ 80%] Meshing curve 160 (Line)
Info : [ 80%] Meshing curve 161 (Line)
Info : [ 80%] Meshing curve 162 (Line)
Info : [ 80%] Meshing curve 163 (Line)
Info : [ 80%] Meshing curve 164 (Line)
Info : [ 80%] Meshing curve 1234 (Line)
Info : [ 90%] Meshing curve 3005 (Circle)
Info : [ 90%] Meshing curve 3006 (Circle)
Info : [ 90%] Meshing curve 3007 (Circle)
Info : [ 90%] Meshing curve 3008 (Circle)
Info : [ 90%] Meshing curve 3015 (Circle)
Info : [ 90%] Meshing curve 3016 (Circle)
Info : [ 90%] Meshing curve 3017 (Circle)
Info : [ 90%] Meshing curve 3018 (Circle)
Info : [100%] Meshing curve 3025 (Circle)
Info : [100%] Meshing curve 3026 (Circle)
Info : [100%] Meshing curve 3027 (Circle)
Info : [100%] Meshing curve 3028 (Circle)
Info : [100%] Meshing curve 3035 (Circle)
Info : [100%] Meshing curve 3036 (Circle)
Info : [100%] Meshing curve 3037 (Circle)
Info : [100%] Meshing curve 3038 (Circle)
Info : Done meshing 1D (Wall 0.0061591s, CPU 0.005577s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 200 (Plane, Delaunay)
Info : [ 0%] Blossom: 8705 internal 168 closed
Info : [ 0%] Blossom recombination completed (Wall 0.0738333s, CPU 0.073509s): 2854 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.769982, min Q = 0.452527
Info : [ 30%] Meshing surface 201 (Plane, Delaunay)
Info : [ 30%] Blossom: 8008 internal 400 closed
Info : [ 30%] Blossom recombination completed (Wall 0.0766879s, CPU 0.06083s): 2590 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.791636, min Q = 0.499656
Info : [ 50%] Meshing surface 202 (Plane, Delaunay)
Info : [ 50%] Blossom: 1168 internal 472 closed
Info : [ 50%] Blossom recombination completed (Wall 0.00581052s, CPU 0.005768s): 468 quads, 0 triangles, 0 invalid quads, 4 quads with Q < 0.1, avg Q = 0.749983, min Q = 0.0166667
Info : [ 70%] Meshing surface 203 (Plane, Delaunay)
Info : [ 70%] Blossom: 1208 internal 488 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00904007s, CPU 0.008077s): 480 quads, 8 triangles, 0 invalid quads, 4 quads with Q < 0.1, avg Q = 0.672659, min Q = 0.016129
Info : [ 90%] Meshing surface 204 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.29722s, CPU 0.257248s)
Info : 6777 nodes 8146 elements
Info : Writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/GreatCell.msh'
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/domain.vtu (7016 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_Fracture.vtu (16 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_Central_sample.vtu (2854 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_OuterPart_sample.vtu (3546 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture/physical_group_Rubber_sheath.vtu (600 cells)

%cd {mesh_path_embedded}…
(click to toggle)
%cd {mesh_path_embedded}
!pwd
run(
"identifySubdomains -f -m domain.vtu -- "
"physical_group_DSS1.vtu physical_group_DSS1a.vtu "
"physical_group_DSS2.vtu physical_group_DSS2a.vtu "
"physical_group_DSS3.vtu physical_group_DSS3a.vtu "
"physical_group_DSS4.vtu physical_group_DSS4a.vtu "
"physical_group_DSS5.vtu physical_group_DSS5a.vtu "
"physical_group_DSS6.vtu physical_group_DSS6a.vtu "
"physical_group_DSS7.vtu physical_group_DSS7a.vtu "
"physical_group_DSS8.vtu physical_group_DSS8a.vtu "
"physical_group_PEE1.vtu physical_group_PEE1a.vtu "
"physical_group_PEE2.vtu physical_group_PEE2a.vtu "
"physical_group_PEE3.vtu physical_group_PEE3a.vtu "
"physical_group_PEE4.vtu physical_group_PEE4a.vtu "
"physical_group_PEE5.vtu physical_group_PEE5a.vtu "
"physical_group_PEE6.vtu physical_group_PEE6a.vtu "
"physical_group_PEE7.vtu physical_group_PEE7a.vtu "
"physical_group_PEE8.vtu physical_group_PEE8a.vtu "
"physical_group_p_bottom.vtu physical_group_p_left.vtu "
"physical_group_p_right.vtu physical_group_p_top.vtu ",
shell=True,
check=True,
)
%cd -/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture
/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture
info: Mesh reading time: 0.0101767 s
info: MeshNodeSearcher construction time: 0.00031763 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.189e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000657073 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.873e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000250279 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.232e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000253573 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.61e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000244159 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.91e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000246843 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.01e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000243037 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000245381 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.41e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000245641 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.11e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000249477 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000241725 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000247354 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000248835 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.21e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000248596 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.11e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000244569 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.21e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000242907 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.112e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000243899 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.71e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000253423 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.211e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000250299 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.502e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000243849 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000245922 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.172e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000255145 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.252e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000227934 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.82e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000243978 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.212e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000244149 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.262e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000252351 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.222e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000250108 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.091e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000253924 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.112e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000243878 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.132e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000252691 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.492e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000244409 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.72e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000242706 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.383e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.0002447 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.41e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_bottom' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.00025133 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 2.91e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_left' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000240954 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 2.7e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_right' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000243738 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 2.61e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_top' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000248345 s
info: identifySubdomains time: 0.00955332 s
info: writing time: 0.0235724 s
info: Entire run time: 0.0441742 s
/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
Run the simulation
Input
# Times for load curves…
(click to toggle)
# Times for load curves
times = "0.0 1000. 3500"
simulation_end_time = 3500.0
n_fracture_p_ncs = 3
model_type = "M2b"
output_prefix = "M2b_LIE"
# Load
PEE_load_values = {
"A": [10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6, 9.95e6],
"B": [7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6],
"C": [1.0e6, 3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6],
}
# Project file
prj_file = Path("M2_LIE.prj")
prj = ot.Project(input_file=prj_file, output_file=Path(out_dir, f"{output_prefix}.prj"))Run OGS
sing_ogs_model = SingleOGSModel(…
(click to toggle)
sing_ogs_model = SingleOGSModel(
model=prj,
out_dir=out_dir,
mesh_path=mesh_path_embedded,
output_prefix=output_prefix,
method="LIE",
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
materials=materials,
)
vtu_files_dict_embedded = sing_ogs_model.run_simulations_with_fracture(
times=times,
base_project_file=prj_file,
mesh_path=mesh_path_embedded,
load_cases=PEE_load_values,
material_names=material_names,
materials=materials,
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
output_prefix=output_prefix,
out_dir=out_dir,
use_b_bar_value="true",
tension_cutoff="true",
)mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2b_LIE.prj
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2b_LIE_A.prj
============================================================
Running simulation for load case: A with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2b_LIE_A.prj
Status: finished successfully.
Execution took 27.00706124305725 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2b_LIE_A.prj
Status: finished successfully.
Execution took 16.285318613052368 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2b_LIE_B.prj
============================================================
Running simulation for load case: B with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2b_LIE_B.prj
Status: finished successfully.
Execution took 17.452633380889893 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2b_LIE_B.prj
Status: finished successfully.
Execution took 20.357427835464478 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_embeddedFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2b_LIE_C.prj
============================================================
Running simulation for load case: C with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2b_LIE_C.prj
Status: finished successfully.
Execution took 25.240665674209595 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2b_LIE_C.prj
Status: finished successfully.
Execution took 30.187623739242554 s
Post-processing
Volumetric strain vs angle at probe circle
plotter = Plotter(…
(click to toggle)
plotter = Plotter(
output_dir=out_dir,
save_extracted_data=True,
)
plotter.plot_volumetric_strain_vs_angle(
vtu_files_dict_embedded,
model_type="M2b",
layout="single",
ylim_range=[-7.5, 2.5],
)

Profiles
plotter.plot_field_variables(vtu_files_dict_embedded)Load: A, File 0, Benchmark: M2b, Material: Gneiss

Load: A, File 1, Benchmark: M2b, Material: Greywacke

Load: B, File 0, Benchmark: M2b, Material: Gneiss

Load: B, File 1, Benchmark: M2b, Material: Greywacke

Load: C, File 0, Benchmark: M2b, Material: Gneiss

Load: C, File 1, Benchmark: M2b, Material: Greywacke

Fully fractured sample ($\texttt{M}_{2a}$)
In this benchmark, 2D plane strain numerical simulations are performed to establish a baseline model for assessing the impact of fracture orientation on strain distribution under poly-axial loading. A planar fracture within the specimen, $\Gamma = \left[-0.094, 0.094\right] \times \{0\}$, is considered, under poly-axial loading applied on PEE’s and DSS’s
Mesh Generation
Input
h = 0.0025…
(click to toggle)
h = 0.0025
meshname = "GreatCell"
mesh_path_full = Path(".", out_dir, "mesh_GreatCell_fullFracture").resolve()Boundary meshes
Gmsh (boundary meshes)
msh_file_full = mesh_GreatCell_fullFracture(…
(click to toggle)
msh_file_full = mesh_GreatCell_fullFracture(
lc=0.005,
lc2=h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path_full,
meshname=meshname,
mode="BC",
post_process=True,
cmap="viridis",
opacity=0.8,
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 20%] Meshing curve 110 (Line)
Info : [ 20%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 20%] Meshing curve 118 (Line)
Info : [ 30%] Meshing curve 119 (Line)
Info : [ 30%] Meshing curve 120 (Line)
Info : [ 30%] Meshing curve 121 (Line)
Info : [ 30%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 30%] Meshing curve 126 (Line)
Info : [ 30%] Meshing curve 127 (Line)
Info : [ 40%] Meshing curve 128 (Line)
Info : [ 40%] Meshing curve 129 (Line)
Info : [ 40%] Meshing curve 130 (Line)
Info : [ 40%] Meshing curve 131 (Line)
Info : [ 40%] Meshing curve 132 (Line)
Info : [ 40%] Meshing curve 133 (Line)
Info : [ 40%] Meshing curve 134 (Line)
Info : [ 40%] Meshing curve 135 (Line)
Info : [ 40%] Meshing curve 136 (Line)
Info : [ 50%] Meshing curve 137 (Line)
Info : [ 50%] Meshing curve 138 (Line)
Info : [ 50%] Meshing curve 139 (Line)
Info : [ 50%] Meshing curve 140 (Line)
Info : [ 50%] Meshing curve 141 (Line)
Info : [ 50%] Meshing curve 142 (Line)
Info : [ 50%] Meshing curve 143 (Line)
Info : [ 50%] Meshing curve 144 (Line)
Info : [ 60%] Meshing curve 145 (Line)
Info : [ 60%] Meshing curve 146 (Line)
Info : [ 60%] Meshing curve 147 (Line)
Info : [ 60%] Meshing curve 148 (Line)
Info : [ 60%] Meshing curve 149 (Line)
Info : [ 60%] Meshing curve 150 (Line)
Info : [ 60%] Meshing curve 151 (Line)
Info : [ 60%] Meshing curve 152 (Line)
Info : [ 60%] Meshing curve 153 (Line)
Info : [ 70%] Meshing curve 154 (Line)
Info : [ 70%] Meshing curve 155 (Line)
Info : [ 70%] Meshing curve 156 (Line)
Info : [ 70%] Meshing curve 157 (Line)
Info : [ 70%] Meshing curve 158 (Line)
Info : [ 70%] Meshing curve 159 (Line)
Info : [ 70%] Meshing curve 160 (Line)
Info : [ 70%] Meshing curve 161 (Line)
Info : [ 70%] Meshing curve 162 (Line)
Info : [ 80%] Meshing curve 163 (Line)
Info : [ 80%] Meshing curve 164 (Line)
Info : [ 80%] Meshing curve 3005 (Circle)
Info : [ 80%] Meshing curve 3006 (Circle)
Info : [ 80%] Meshing curve 3007 (Circle)
Info : [ 80%] Meshing curve 3008 (Circle)
Info : [ 80%] Meshing curve 3015 (Circle)
Info : [ 80%] Meshing curve 3016 (Circle)
Info : [ 80%] Meshing curve 3017 (Circle)
Info : [ 90%] Meshing curve 3018 (Circle)
Info : [ 90%] Meshing curve 3025 (Circle)
Info : [ 90%] Meshing curve 3026 (Circle)
Info : [ 90%] Meshing curve 3027 (Circle)
Info : [ 90%] Meshing curve 3028 (Circle)
Info : [ 90%] Meshing curve 3035 (Circle)
Info : [ 90%] Meshing curve 3036 (Circle)
Info : [ 90%] Meshing curve 3037 (Circle)
Info : [ 90%] Meshing curve 3038 (Circle)
Info : [100%] Meshing curve 8000 (Line)
Info : [100%] Meshing curve 8001 (Line)
Info : [100%] Meshing curve 8002 (Line)
Info : [100%] Meshing curve 8003 (Line)
Info : [100%] Meshing curve 8004 (Line)
Info : [100%] Meshing curve 8005 (Line)
Info : [100%] Meshing curve 8006 (Line)
Info : [100%] Meshing curve 8007 (Line)
Info : Done meshing 1D (Wall 0.00588627s, CPU 0.006035s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 200 (Plane, Delaunay)
Info : [ 0%] Blossom: 4270 internal 134 closed
Info : [ 0%] Blossom recombination completed (Wall 0.0362775s, CPU 0.036066s): 1404 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.768417, min Q = 0.451439
Info : [ 20%] Meshing surface 201 (Plane, Delaunay)
Info : [ 20%] Blossom: 3961 internal 220 closed
Info : [ 20%] Blossom recombination completed (Wall 0.0332476s, CPU 0.033099s): 1302 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.77867, min Q = 0.477434
Info : [ 30%] Meshing surface 202 (Plane, Delaunay)
Info : [ 30%] Blossom: 591 internal 240 closed
Info : [ 30%] Blossom recombination completed (Wall 0.00316955s, CPU 0.003141s): 235 quads, 0 triangles, 0 invalid quads, 2 quads with Q < 0.1, avg Q = 0.762906, min Q = 0.0166667
Info : [ 40%] Meshing surface 203 (Plane, Delaunay)
Info : [ 40%] Blossom: 599 internal 244 closed
Info : [ 40%] Blossom recombination completed (Wall 0.00455s, CPU 0.004514s): 238 quads, 6 triangles, 0 invalid quads, 3 quads with Q < 0.1, avg Q = 0.630126, min Q = 0.016129
Info : [ 50%] Meshing surface 204 (Plane, Delaunay)
Info : [ 50%] Blossom: 4270 internal 134 closed
Info : [ 50%] Blossom recombination completed (Wall 0.0366392s, CPU 0.036464s): 1404 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.768413, min Q = 0.451439
Info : [ 60%] Meshing surface 205 (Plane, Delaunay)
Info : [ 60%] Blossom: 3961 internal 220 closed
Info : [ 60%] Blossom recombination completed (Wall 0.0330883s, CPU 0.033011s): 1302 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.77867, min Q = 0.477434
Info : [ 70%] Meshing surface 206 (Plane, Delaunay)
Info : [ 70%] Blossom: 591 internal 240 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00333497s, CPU 0.003301s): 235 quads, 0 triangles, 0 invalid quads, 2 quads with Q < 0.1, avg Q = 0.762906, min Q = 0.0166667
Info : [ 80%] Meshing surface 207 (Plane, Delaunay)
Info : [ 80%] Blossom: 599 internal 244 closed
Info : [ 80%] Blossom recombination completed (Wall 0.00465865s, CPU 0.004622s): 238 quads, 6 triangles, 0 invalid quads, 3 quads with Q < 0.1, avg Q = 0.630126, min Q = 0.016129
Info : [ 90%] Meshing surface 208 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.262099s, CPU 0.260384s)
Info : 6745 nodes 8179 elements
Info : Writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/GreatCell.msh'
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/domain.vtu (160 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_Inlet.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_p_right.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_p_top.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_p_left.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_p_bottom.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_Outlet_R.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_Outlet_L.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE1.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE2.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE3.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE4.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE5.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE6.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE7.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE8.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE1a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE2a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE3a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE4a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE5a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE6a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE7a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_PEE8a.vtu (6 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS1.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS2.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS3.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS4.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS5.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS6.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS7.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS8.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS1a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS2a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS3a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS4a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS5a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS6a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS7a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_DSS8a.vtu (4 cells)

Computational domain mesh
Gmsh (computational domain mesh)
msh_file_full = mesh_GreatCell_fullFracture(…
(click to toggle)
msh_file_full = mesh_GreatCell_fullFracture(
lc=0.005,
lc2=h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path_full,
meshname=meshname,
mode="domain",
post_process=True,
cmap="viridis",
opacity=0.8,
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 20%] Meshing curve 110 (Line)
Info : [ 20%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 20%] Meshing curve 118 (Line)
Info : [ 30%] Meshing curve 119 (Line)
Info : [ 30%] Meshing curve 120 (Line)
Info : [ 30%] Meshing curve 121 (Line)
Info : [ 30%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 30%] Meshing curve 126 (Line)
Info : [ 30%] Meshing curve 127 (Line)
Info : [ 40%] Meshing curve 128 (Line)
Info : [ 40%] Meshing curve 129 (Line)
Info : [ 40%] Meshing curve 130 (Line)
Info : [ 40%] Meshing curve 131 (Line)
Info : [ 40%] Meshing curve 132 (Line)
Info : [ 40%] Meshing curve 133 (Line)
Info : [ 40%] Meshing curve 134 (Line)
Info : [ 40%] Meshing curve 135 (Line)
Info : [ 40%] Meshing curve 136 (Line)
Info : [ 50%] Meshing curve 137 (Line)
Info : [ 50%] Meshing curve 138 (Line)
Info : [ 50%] Meshing curve 139 (Line)
Info : [ 50%] Meshing curve 140 (Line)
Info : [ 50%] Meshing curve 141 (Line)
Info : [ 50%] Meshing curve 142 (Line)
Info : [ 50%] Meshing curve 143 (Line)
Info : [ 50%] Meshing curve 144 (Line)
Info : [ 60%] Meshing curve 145 (Line)
Info : [ 60%] Meshing curve 146 (Line)
Info : [ 60%] Meshing curve 147 (Line)
Info : [ 60%] Meshing curve 148 (Line)
Info : [ 60%] Meshing curve 149 (Line)
Info : [ 60%] Meshing curve 150 (Line)
Info : [ 60%] Meshing curve 151 (Line)
Info : [ 60%] Meshing curve 152 (Line)
Info : [ 60%] Meshing curve 153 (Line)
Info : [ 70%] Meshing curve 154 (Line)
Info : [ 70%] Meshing curve 155 (Line)
Info : [ 70%] Meshing curve 156 (Line)
Info : [ 70%] Meshing curve 157 (Line)
Info : [ 70%] Meshing curve 158 (Line)
Info : [ 70%] Meshing curve 159 (Line)
Info : [ 70%] Meshing curve 160 (Line)
Info : [ 70%] Meshing curve 161 (Line)
Info : [ 70%] Meshing curve 162 (Line)
Info : [ 80%] Meshing curve 163 (Line)
Info : [ 80%] Meshing curve 164 (Line)
Info : [ 80%] Meshing curve 3005 (Circle)
Info : [ 80%] Meshing curve 3006 (Circle)
Info : [ 80%] Meshing curve 3007 (Circle)
Info : [ 80%] Meshing curve 3008 (Circle)
Info : [ 80%] Meshing curve 3015 (Circle)
Info : [ 80%] Meshing curve 3016 (Circle)
Info : [ 80%] Meshing curve 3017 (Circle)
Info : [ 90%] Meshing curve 3018 (Circle)
Info : [ 90%] Meshing curve 3025 (Circle)
Info : [ 90%] Meshing curve 3026 (Circle)
Info : [ 90%] Meshing curve 3027 (Circle)
Info : [ 90%] Meshing curve 3028 (Circle)
Info : [ 90%] Meshing curve 3035 (Circle)
Info : [ 90%] Meshing curve 3036 (Circle)
Info : [ 90%] Meshing curve 3037 (Circle)
Info : [ 90%] Meshing curve 3038 (Circle)
Info : [100%] Meshing curve 8000 (Line)
Info : [100%] Meshing curve 8001 (Line)
Info : [100%] Meshing curve 8002 (Line)
Info : [100%] Meshing curve 8003 (Line)
Info : [100%] Meshing curve 8004 (Line)
Info : [100%] Meshing curve 8005 (Line)
Info : [100%] Meshing curve 8006 (Line)
Info : [100%] Meshing curve 8007 (Line)
Info : Done meshing 1D (Wall 0.00546435s, CPU 0.005532s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 200 (Plane, Delaunay)
Info : [ 0%] Blossom: 4270 internal 134 closed
Info : [ 0%] Blossom recombination completed (Wall 0.0349493s, CPU 0.034757s): 1404 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.768417, min Q = 0.451439
Info : [ 20%] Meshing surface 201 (Plane, Delaunay)
Info : [ 20%] Blossom: 3961 internal 220 closed
Info : [ 20%] Blossom recombination completed (Wall 0.0313649s, CPU 0.031224s): 1302 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.77867, min Q = 0.477434
Info : [ 30%] Meshing surface 202 (Plane, Delaunay)
Info : [ 30%] Blossom: 591 internal 240 closed
Info : [ 30%] Blossom recombination completed (Wall 0.00304135s, CPU 0.003026s): 235 quads, 0 triangles, 0 invalid quads, 2 quads with Q < 0.1, avg Q = 0.762906, min Q = 0.0166667
Info : [ 40%] Meshing surface 203 (Plane, Delaunay)
Info : [ 40%] Blossom: 599 internal 244 closed
Info : [ 40%] Blossom recombination completed (Wall 0.00437786s, CPU 0.004358s): 238 quads, 6 triangles, 0 invalid quads, 3 quads with Q < 0.1, avg Q = 0.630126, min Q = 0.016129
Info : [ 50%] Meshing surface 204 (Plane, Delaunay)
Info : [ 50%] Blossom: 4270 internal 134 closed
Info : [ 50%] Blossom recombination completed (Wall 0.0345571s, CPU 0.034392s): 1404 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.768413, min Q = 0.451439
Info : [ 60%] Meshing surface 205 (Plane, Delaunay)
Info : [ 60%] Blossom: 3961 internal 220 closed
Info : [ 60%] Blossom recombination completed (Wall 0.032213s, CPU 0.030662s): 1302 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.77867, min Q = 0.477434
Info : [ 70%] Meshing surface 206 (Plane, Delaunay)
Info : [ 70%] Blossom: 591 internal 240 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00428385s, CPU 0.003543s): 235 quads, 0 triangles, 0 invalid quads, 2 quads with Q < 0.1, avg Q = 0.762906, min Q = 0.0166667
Info : [ 80%] Meshing surface 207 (Plane, Delaunay)
Info : [ 80%] Blossom: 599 internal 244 closed
Info : [ 80%] Blossom recombination completed (Wall 0.0047484s, CPU 0.004567s): 238 quads, 6 triangles, 0 invalid quads, 3 quads with Q < 0.1, avg Q = 0.630126, min Q = 0.016129
Info : [ 90%] Meshing surface 208 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.256055s, CPU 0.249339s)
Info : 6745 nodes 8179 elements
Info : Writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/GreatCell.msh'
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/domain.vtu (7046 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_fracture.vtu (76 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_Central_sample.vtu (2808 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_OuterPart_sample.vtu (3562 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture/physical_group_Rubber_sheath.vtu (600 cells)

%cd {mesh_path_full}…
(click to toggle)
%cd {mesh_path_full}
run(
"identifySubdomains -f -m domain.vtu -- "
"physical_group_DSS1.vtu physical_group_DSS1a.vtu "
"physical_group_DSS2.vtu physical_group_DSS2a.vtu "
"physical_group_DSS3.vtu physical_group_DSS3a.vtu "
"physical_group_DSS4.vtu physical_group_DSS4a.vtu "
"physical_group_DSS5.vtu physical_group_DSS5a.vtu "
"physical_group_DSS6.vtu physical_group_DSS6a.vtu "
"physical_group_DSS7.vtu physical_group_DSS7a.vtu "
"physical_group_DSS8.vtu physical_group_DSS8a.vtu "
"physical_group_PEE1.vtu physical_group_PEE1a.vtu "
"physical_group_PEE2.vtu physical_group_PEE2a.vtu "
"physical_group_PEE3.vtu physical_group_PEE3a.vtu "
"physical_group_PEE4.vtu physical_group_PEE4a.vtu "
"physical_group_PEE5.vtu physical_group_PEE5a.vtu "
"physical_group_PEE6.vtu physical_group_PEE6a.vtu "
"physical_group_PEE7.vtu physical_group_PEE7a.vtu "
"physical_group_PEE8.vtu physical_group_PEE8a.vtu "
"physical_group_p_bottom.vtu physical_group_p_left.vtu "
"physical_group_p_right.vtu physical_group_p_top.vtu ",
shell=True,
check=True,
)
%cd -/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture
info: Mesh reading time: 0.0109617 s
info: MeshNodeSearcher construction time: 0.000316027 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.617e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000560918 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.012e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000256177 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.11e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000239702 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.11e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.00023811 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.72e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000232702 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.032e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000237659 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.82e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000236587 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000233453 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.01e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000235796 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.01e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000242637 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.41e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000241475 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000239842 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.01e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000240033 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.82e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000252131 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.41e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000237889 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.21e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000239531 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.252e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000236427 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.042e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000241544 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.62e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000241345 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.42e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000244189 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.31e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000240674 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.021e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000247073 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.121e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000240133 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.11e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000242126 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 7.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000243468 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.092e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000245401 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.22e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000245331 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.51e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000237609 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.112e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000241615 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.242e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000242526 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.302e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000243628 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.071e-06 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000244389 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.71e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_bottom' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000244219 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.81e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_left' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000244239 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.2e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_right' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000239131 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 2.8e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_top' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000241955 s
info: identifySubdomains time: 0.00928077 s
info: writing time: 0.0226619 s
info: Entire run time: 0.0435553 s
/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
Run the simulation
Input
# Times for load curves…
(click to toggle)
# Times for load curves
times = "0.0 1000. 3500"
simulation_end_time = 3500.0
n_fracture_p_ncs = 3
model_type = "M2a"
output_prefix = "M2a_LIE"
# Load
PEE_load_values = {
"A": [10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6, 9.95e6],
"B": [7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6],
"C": [1.0e6, 3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6],
}
# Project file
prj_file = Path("M2_LIE.prj")
prj = ot.Project(input_file=prj_file, output_file=Path(out_dir, f"{output_prefix}.prj"))Run OGS
# Create SingleOGSModel…
(click to toggle)
# Create SingleOGSModel
sing_ogs_model = SingleOGSModel(
model=prj,
out_dir=out_dir,
mesh_path=mesh_path_full,
output_prefix=output_prefix,
method="LIE",
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
materials=materials,
)
# Run simulations
vtu_files_dict_full = sing_ogs_model.run_simulations_with_fracture(
times=times,
base_project_file=prj_file,
mesh_path=mesh_path_full,
load_cases=PEE_load_values,
material_names=material_names,
materials=materials,
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
output_prefix=output_prefix,
out_dir=out_dir,
use_b_bar_value="true",
tension_cutoff="true",
)mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2a_LIE.prj
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2a_LIE_A.prj
============================================================
Running simulation for load case: A with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2a_LIE_A.prj
Status: finished successfully.
Execution took 22.02723717689514 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2a_LIE_A.prj
Status: finished successfully.
Execution took 43.49277114868164 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2a_LIE_B.prj
============================================================
Running simulation for load case: B with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2a_LIE_B.prj
Status: finished successfully.
Execution took 21.605637073516846 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2a_LIE_B.prj
Status: finished successfully.
Execution took 31.543155193328857 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/mesh_GreatCell_fullFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2a_LIE_C.prj
============================================================
Running simulation for load case: C with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2a_LIE_C.prj
Status: finished successfully.
Execution took 51.36477613449097 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/Mechanics/GreatCelljupyterNotebook/GreatCellM/M2a_LIE_C.prj
Status: finished successfully.
Execution took 59.08637046813965 s
Post-processing
Volumetric strain vs angle at probe circle
plotter = Plotter(…
(click to toggle)
plotter = Plotter(
output_dir=out_dir,
save_extracted_data=True,
)
plotter.plot_volumetric_strain_vs_angle(
vtu_files_dict_full,
model_type="M2a",
layout="subplots",
ylim_range=[-7.5, 2.5],
)

Profiles
plotter.plot_field_variables(vtu_files_dict_full)Load: A, File 0, Benchmark: M2a, Material: Gneiss

Load: A, File 1, Benchmark: M2a, Material: Greywacke

Load: B, File 0, Benchmark: M2a, Material: Gneiss

Load: B, File 1, Benchmark: M2a, Material: Greywacke

Load: C, File 0, Benchmark: M2a, Material: Gneiss

Load: C, File 1, Benchmark: M2a, Material: Greywacke

pairs_to_check = {…
(click to toggle)
pairs_to_check = {
"M1_SD_A_Greywacke": "M1",
"M2a_LIE_A_Greywacke": "M2a",
"M2b_LIE_A_Greywacke": "M2b",
}
for case, label in pairs_to_check.items():
print(f"\n===== {label} case =====")
new_result = np.load(Path(out_dir, f"extracted_{case}_volStrain.npz"))
expected_result = np.load(Path("expected", f"extracted_{case}_volStrain.npz"))
eps_v_new = new_result["eps_v"]
eps_v_expected = expected_result["eps_v"]
phi_new = new_result["phi"]
phi_expected = expected_result["phi"]
np.testing.assert_allclose(eps_v_new, eps_v_expected, atol=1e-3)
np.testing.assert_allclose(phi_new, phi_expected, atol=1e-8)
print(f"\n{label} case passed.")===== M1 case =====
M1 case passed.
===== M2a case =====
M2a case passed.
===== M2b case =====
M2b case passed.
Reference
-
McDermott, C.I., Fraser-Harris, A., Sauter, M., Couples, G.D., Edlmann, K., Kolditz, O., Lightbody, A., Somerville, J. and Wang, W., 2018. New experimental equipment recreating geo-reservoir conditions in large, fractured, porous samples to investigate coupled thermal, hydraulic and polyaxial stress processes. Scientific reports, 8(1), p.14549.
-
Mollaali, M., Kolditz, O., Hu, M., Park, C.H., Park, J.W., McDermott, C.I., Chittenden, N., Bond, A., Yoon, J.S., Zhou, J. and Pan, P.Z., Liu H., Hou W., Lei H., Zhang L., Nagel T., Barsch M., Wang W., Nguyen S., Kwon S. and Yoshioka K., 2023. Comparative verification of hydro-mechanical fracture behavior: Task G of international research project DECOVALEX–2023. International Journal of Rock Mechanics and Mining Sciences, 170, p.105530.
-
Mollaali, M., Wang, W., You, T., Nagel, T., Fraser-Harris, A., McDermott, C., and Kolditz, O., 2025. Numerical benchmarking of GREAT cell experiments: Poly-axial stress effects on fluid flow in fractured rock using smeared and discrete methods.
This article was written by Mostafa Mollaali, Wenqing Wang. If you are missing something or you find an error please let us know.
Generated with Hugo 0.147.9
in CI job 635984
|
Last revision: April 3, 2025