A 2D GREAT cell benchmark suite simulated using hydro-mechanical and LIE hydro-mechanical processes
import os…
(click to toggle)
import os
import sys
from pathlib import Path
from subprocess import run
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import ogstools as ot
from matplotlib import colormaps
mechanics_path = Path("..", "..", "Mechanics", "GreatCelljupyterNotebook").resolve()
sys.path.insert(0, str(mechanics_path))
from mesh_generator import ( # noqa: E402
mesh_GreatCell_embeddedFracture,
mesh_GreatCell_fullFracture,
mesh_GreatCell_intact,
)
from ogs_model_runner import SingleOGSModel # noqa: E402
from Plotter import Plotter # noqa: E402
def truncated_cmap(name, minval=0.3, maxval=0.9, n=6):…
(click to toggle)
def truncated_cmap(name, minval=0.3, maxval=0.9, n=6):
base = colormaps.get_cmap(name)
return lambda i: base(minval + (maxval - minval) * i / (n - 1))
mpl.rcdefaults()
mpl.rcParams.update(
{
"text.usetex": False,
"font.family": "serif",
"font.serif": ["DejaVu Serif"],
"axes.labelsize": 26,
"axes.titlesize": 24,
"legend.fontsize": 18,
"xtick.labelsize": 18,
"ytick.labelsize": 18,
}
)
ot.plot.setup.show_region_bounds = False…
(click to toggle)
ot.plot.setup.show_region_bounds = False
out_dir = Path(os.environ.get("OGS_TESTRUNNER_OUT_DIR", "_out"))
out_dir.mkdir(parents=True, exist_ok=True)Great cell
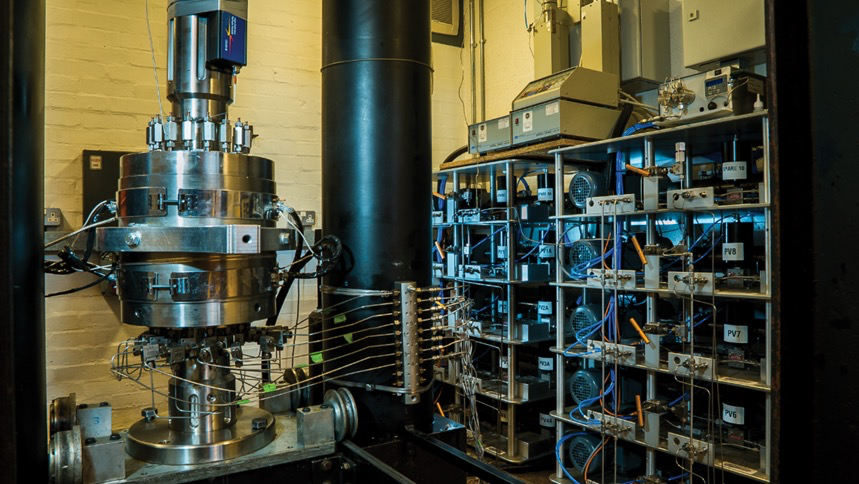
The GREAT cell is a poly-axial rock-testing device that reproduces subsurface conditions down to 3.5 km depth on 200 mm-diameter samples. It imposes a rotating stress field, injects fluid through a central borehole, and records both fiber-optic strain and pore-pressure data—providing a rich dataset for validating coupled hydro-mechanical models. For full details, see the GREAT cell benchmark docs: www.opengeosys.org/docs/benchmarks/small-deformations/greatcellm/
Here, in our 2D, small-deformation benchmark suite, we employ the LIE (lower-dimensional interface element) hydro-mechanical process to model fractures as interfaces. The cases are:
- $\texttt{HM}_1$: Intact rock sample with fluid injection.
- $\texttt{HM}_{2a}$: Fully fractured sample with inflow-outflow.
- $\texttt{HM}_{2b}$: Half-fractured sample with inflow-outflow.
Material Properties
materials = {…
(click to toggle)
materials = {
"Gneiss": {
"young_sample": 83.9e9, # Young's modulus (Pa)
"nu_sample": 0.21, # Poisson's ratio
"biot": 0.6, # Biot coefficient
"porosity": 0.001, # Porosity
"permeability": 1e-19, # Permeability (m²)
"density_solid": 2750, # Solid density (kg/m³)
"k_n": 200e9, # Normal stiffness (Pa/m)
"k_t": 100e9, # Tangential stiffness (Pa/m)
"c_f": 4.4e-10, # Fluid compressibility (Pa⁻¹)
"k_s": 4.82e10, # Solid bulk modulus (Pa)
"S_f": 4.4e-10, # Specific storage (Pa⁻¹)
"t_np": 10e6, # Peak normal traction (Pa)
"Gc": 50, # Fracture toughness (J/m²)
"w_init": 1e-6, # initial fracture width (m)
"fluid": {
"density": 1000.0, # Fluid density (kg/m³)
"viscosity": 1e-3, # Fluid viscosity (Pa·s)
"injectionFlowRate_Inlet": 4.167e-7, # Injection flow rate (m³/s)
"p_outlet": 3.45e6, # Outlet pressure (Pa)
},
"rubber_sheath": {
"young_modulus": 0.1e9, # Young's modulus (Pa)
"poisson_ratio": 0.4, # Poisson's ratio
"porosity": 0.001, # Porosity
"permeability": 1e-17, # Permeability (m²)
"density": 1500, # Density (kg/m³)
"biot": 0.0, # Biot coefficient
},
},
"Greywacke": {
"young_sample": 26.87e9, # Young's modulus (Pa)
"nu_sample": 0.27, # Poisson's ratio
"biot": 0.8, # Biot coefficient
"porosity": 0.005, # Porosity
"permeability": 2.58e-19, # Permeability (m²)
"density_solid": 2650, # Solid density (kg/m³)
"k_n": 100e9, # Normal stiffness (Pa/m)
"k_t": 50e9, # Tangential stiffness (Pa/m)
"c_f": 4.4e-10, # Fluid compressibility (Pa⁻¹)
"k_s": 1.95e10, # Solid bulk modulus (Pa)
"S_f": 4.4e-10, # Specific storage (Pa⁻¹)
"t_np": 10e6, # Peak normal traction (Pa)
"Gc": 30, # Fracture toughness (J/m²)
"w_init": 1e-6, # initial fracture width (m)
"fluid": {
"density": 1000.0, # Fluid density (kg/m³)
"viscosity": 1e-3, # Fluid viscosity (Pa·s)
"injectionFlowRate_Inlet": 4.167e-7, # Injection flow rate (m³/s)
"p_outlet": 3.45e6, # Outlet pressure (Pa)
},
"rubber_sheath": {
"young_modulus": 0.1e9, # Young's modulus (Pa)
"poisson_ratio": 0.4, # Poisson's ratio
"porosity": 0.001, # Porosity
"permeability": 1e-17, # Permeability (m²)
"density": 1500, # Density (kg/m³)
"biot": 0.0, # Biot coefficient
},
},
}
material_names = list(materials.keys())Loads
Table: Load Conditions
| Marker | PEE1 Angle to $\sigma_2$ | PEE1 & 1a | PEE2 & 2a | PEE3 & 3a | PEE4 & 4a | PEE5 & 5a | PEE6 & 6a | PEE7 & 7a | PEE8 & 8a |
|---|---|---|---|---|---|---|---|---|---|
| A | 0° | 10.0 | 6.64 | 4.46 | 1.17 | 1.0 | 3.82 | 7.80 | 9.95 |
| E | 22.5° | 9.95 | 10.0 | 6.64 | 4.46 | 1.17 | 1.0 | 3.82 | 7.80 |
| B | 45.0° | 7.80 | 9.95 | 10.0 | 6.64 | 4.46 | 1.17 | 1.0 | 3.82 |
| F | 67.5° | 3.82 | 7.80 | 9.95 | 10.0 | 6.64 | 4.46 | 1.17 | 1.0 |
| C | 90° | 1.0 | 3.82 | 7.80 | 9.95 | 10.0 | 6.64 | 4.46 | 1.17 |
All loads are in MPa. DSS loads are averages of adjacent PEEs.
Schematic loading conditions
loads = {…
(click to toggle)
loads = {
"A": [
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
7.80e6,
9.95e6,
],
"B": [
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
],
"C": [
1.0e6,
3.82e6,
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
1.0e6,
3.82e6,
7.80e6,
9.95e6,
10.0e6,
6.64e6,
4.46e6,
1.17e6,
],
}
angles_degrees = […
(click to toggle)
angles_degrees = [
0,
-22.5,
-45,
-67.5,
-90,
-112.5,
-135,
-157.5,
-180,
-202.5,
-225,
-247.5,
-270,
-292.5,
-315,
-337.5,
]
angles_radians = np.deg2rad(angles_degrees)
circle_radius = 6
circle_x = circle_radius * np.cos(angles_radians)
circle_y = circle_radius * np.sin(angles_radians)
fig, axs = plt.subplots(1, 3, figsize=(21, 7), facecolor="none")
for idx, (label, values) in enumerate(loads.items()):
ax = axs[idx]
ax.set_aspect("equal")
ax.axis("off")
formatted_values = [rf"${v / 1e6:.1f}$" for v in values]
scaled_values = [v / 2 for v in values]
circle = plt.Circle(
(0, 0),
circle_radius,
color="black",
fill=False,
linestyle="--",
linewidth=2,
)
ax.add_artist(circle)
top_points_x, top_points_y = [], []
for i, (x, y, value) in enumerate(
zip(circle_x, circle_y, scaled_values, strict=True)
):
unit_vector = np.array([x, y]) / circle_radius
line_end = np.array([x, y]) + unit_vector * value / 1e6
top_points_x.append(line_end[0])
top_points_y.append(line_end[1])
ax.annotate(
"",
xytext=line_end,
xy=(x, y),
arrowprops={
"arrowstyle": "-|>",
"color": "blue",
"lw": 2,
"mutation_scale": 15,
"fill": True,
},
)
angle_offset = 1.2 * unit_vector
angle_label = f"{-angles_degrees[i]}°"
ax.text(
x - angle_offset[0],
y - angle_offset[1],
angle_label,
fontsize=12,
ha="center",
va="center",
color="m",
)
value_offset = 0.5 * unit_vector
ax.text(
line_end[0] + 3.0 * value_offset[0],
line_end[1] + 1.8 * value_offset[1],
formatted_values[i],
fontsize=18,
ha="center",
weight="bold",
)
top_points_x.append(top_points_x[0])
top_points_y.append(top_points_y[0])
ax.plot(
top_points_x,
top_points_y,
color="green",
linestyle="-.",
lw=2,
marker="o",
markersize=6,
markerfacecolor="lightgreen",
markeredgewidth=1.0,
markeredgecolor="black",
)
ax.set_xlim([-12, 12])
ax.set_ylim([-12, 12])
ax.text(
0,
0,
f"Load {label}",
fontsize=32,
ha="center",
va="center",
family="serif",
)
plt.tight_layout()
output_path = Path(out_dir, "loads_A_B_C_schematic_with_angles.png")
plt.savefig(output_path, dpi=350, bbox_inches="tight", transparent=True)
plt.show()

Boundary Conditions
The boundary conditions applied in the simulation include both Dirichlet and Neumann conditions.
-
Dirichlet conditions (displacement control): \begin{equation*} \begin{cases} u_x(0, y) = 0, \quad u_y(0, y) = 0 & \quad \text{for } y = -0.09894 \text{ m}, \ u_y(x, 0) = 0, & \quad \text{for} x = -0.09894 \text{ m}. \end{cases} \end{equation*}
-
Neumann conditions:
Normal stress $\sigma_{rr}$ is applied on PEEs and DSSs. The DSS stress is calculated as:
$$\begin{equation*}\sigma_\text{DSS}^i = \frac{\sigma_\text{PEE}^i + \sigma_\text{PEE}^{i+1}}{2}\end{equation*} $$
Intact rock sample with fluid injection ($\texttt{HM}_1$)
This benchmark, which does not consider any fracture, is designed to verify the basic computational setting for hydro-mechanical simulations. In addition to the mechanical loads, a zero constant pore pressure is prescribed at the outer boundary. {At the center of sample, fluid is injected at a rate of $Q_0^{\text{v}} = 2.085 \times 10^{-9}$~m$^3$/s,}$
The hydro-mechanical simulations follows a two-stage process: a 3000s equilibrium phase under mechanical loading to stabilize initial conditions, followed by a 500s fluid injection phase to model fluid flow. This loading condition is applied to both Greywacke and Gneiss samples
Mesh Generation
Input
h = 0.005…
(click to toggle)
h = 0.005
meshname = "GreatCell"
mesh_path = Path(out_dir, "mesh_intact").resolve()
print(mesh_path)/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact
Boundary meshes
Gmsh (boundary meshes)
msh_file = mesh_GreatCell_intact(…
(click to toggle)
msh_file = mesh_GreatCell_intact(
lc=2 * h,
lc2=h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path,
meshname=meshname,
mode="BC",
post_process=True,
cmap="viridis",
opacity=0.8,
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 20%] Meshing curve 110 (Line)
Info : [ 20%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 30%] Meshing curve 118 (Line)
Info : [ 30%] Meshing curve 119 (Line)
Info : [ 30%] Meshing curve 120 (Line)
Info : [ 30%] Meshing curve 121 (Line)
Info : [ 30%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 40%] Meshing curve 126 (Line)
Info : [ 40%] Meshing curve 127 (Line)
Info : [ 40%] Meshing curve 128 (Line)
Info : [ 40%] Meshing curve 129 (Line)
Info : [ 40%] Meshing curve 130 (Line)
Info : [ 40%] Meshing curve 131 (Line)
Info : [ 40%] Meshing curve 132 (Line)
Info : [ 40%] Meshing curve 133 (Line)
Info : [ 50%] Meshing curve 134 (Line)
Info : [ 50%] Meshing curve 135 (Line)
Info : [ 50%] Meshing curve 136 (Line)
Info : [ 50%] Meshing curve 137 (Line)
Info : [ 50%] Meshing curve 138 (Line)
Info : [ 50%] Meshing curve 139 (Line)
Info : [ 50%] Meshing curve 140 (Line)
Info : [ 50%] Meshing curve 141 (Line)
Info : [ 60%] Meshing curve 142 (Line)
Info : [ 60%] Meshing curve 143 (Line)
Info : [ 60%] Meshing curve 144 (Line)
Info : [ 60%] Meshing curve 145 (Line)
Info : [ 60%] Meshing curve 146 (Line)
Info : [ 60%] Meshing curve 147 (Line)
Info : [ 60%] Meshing curve 148 (Line)
Info : [ 60%] Meshing curve 149 (Line)
Info : [ 70%] Meshing curve 150 (Line)
Info : [ 70%] Meshing curve 151 (Line)
Info : [ 70%] Meshing curve 152 (Line)
Info : [ 70%] Meshing curve 153 (Line)
Info : [ 70%] Meshing curve 154 (Line)
Info : [ 70%] Meshing curve 155 (Line)
Info : [ 70%] Meshing curve 156 (Line)
Info : [ 70%] Meshing curve 157 (Line)
Info : [ 80%] Meshing curve 158 (Line)
Info : [ 80%] Meshing curve 159 (Line)
Info : [ 80%] Meshing curve 160 (Line)
Info : [ 80%] Meshing curve 161 (Line)
Info : [ 80%] Meshing curve 162 (Line)
Info : [ 80%] Meshing curve 163 (Line)
Info : [ 80%] Meshing curve 164 (Line)
Info : [ 80%] Meshing curve 1234 (Line)
Info : [ 90%] Meshing curve 3005 (Circle)
Info : [ 90%] Meshing curve 3006 (Circle)
Info : [ 90%] Meshing curve 3007 (Circle)
Info : [ 90%] Meshing curve 3008 (Circle)
Info : [ 90%] Meshing curve 3015 (Circle)
Info : [ 90%] Meshing curve 3016 (Circle)
Info : [ 90%] Meshing curve 3017 (Circle)
Info : [ 90%] Meshing curve 3018 (Circle)
Info : [100%] Meshing curve 3025 (Circle)
Info : [100%] Meshing curve 3026 (Circle)
Info : [100%] Meshing curve 3027 (Circle)
Info : [100%] Meshing curve 3028 (Circle)
Info : [100%] Meshing curve 3035 (Circle)
Info : [100%] Meshing curve 3036 (Circle)
Info : [100%] Meshing curve 3037 (Circle)
Info : [100%] Meshing curve 3038 (Circle)
Info : Done meshing 1D (Wall 0.00906916s, CPU 0.00482s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 200 (Plane, Delaunay)
Info : [ 0%] Blossom: 2264 internal 88 closed
Info : [ 0%] Blossom recombination completed (Wall 0.0159058s, CPU 0.015793s): 740 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.776443, min Q = 0.460287
Info : [ 30%] Meshing surface 201 (Plane, Delaunay)
Info : [ 30%] Blossom: 2128 internal 208 closed
Info : [ 30%] Blossom recombination completed (Wall 0.0130688s, CPU 0.01302s): 744 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.715401, min Q = 0.462407
Info : [ 50%] Meshing surface 202 (Plane, Delaunay)
Info : [ 50%] Blossom: 600 internal 240 closed
Info : [ 50%] Blossom recombination completed (Wall 0.00224414s, CPU 0.00221s): 120 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.983333, min Q = 0.983333
Info : [ 70%] Meshing surface 203 (Plane, Delaunay)
Info : [ 70%] Blossom: 344 internal 248 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00188152s, CPU 0.001874s): 132 quads, 16 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.59063, min Q = 0.274456
Info : [ 90%] Meshing surface 204 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.0691628s, CPU 0.064425s)
Info : 1937 nodes 2682 elements
Info : Writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/GreatCell.msh'
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/domain.vtu (96 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_Inlet.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_p_right.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_p_top.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_p_left.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_p_bottom.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE1.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE2.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE3.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE4.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE5.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE6.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE7.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE8.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE1a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE2a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE3a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE4a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE5a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE6a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE7a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_PEE8a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS1.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS2.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS3.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS4.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS5.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS6.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS7.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS8.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS1a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS2a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS3a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS4a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS5a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS6a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS7a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_DSS8a.vtu (2 cells)
[0m[33m2025-10-23 14:27:23.983 ( 3.100s) [ 7F02FD01A400]vtkXOpenGLRenderWindow.:1416 WARN| bad X server connection. DISPLAY=[0m

Computational domain mesh
Gmsh
msh_file = mesh_GreatCell_intact(…
(click to toggle)
msh_file = mesh_GreatCell_intact(
lc=2 * h,
lc2=h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path,
meshname=meshname,
mode="domain",
post_process=True,
cmap="viridis",
opacity=0.8,
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 20%] Meshing curve 110 (Line)
Info : [ 20%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 30%] Meshing curve 118 (Line)
Info : [ 30%] Meshing curve 119 (Line)
Info : [ 30%] Meshing curve 120 (Line)
Info : [ 30%] Meshing curve 121 (Line)
Info : [ 30%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 40%] Meshing curve 126 (Line)
Info : [ 40%] Meshing curve 127 (Line)
Info : [ 40%] Meshing curve 128 (Line)
Info : [ 40%] Meshing curve 129 (Line)
Info : [ 40%] Meshing curve 130 (Line)
Info : [ 40%] Meshing curve 131 (Line)
Info : [ 40%] Meshing curve 132 (Line)
Info : [ 40%] Meshing curve 133 (Line)
Info : [ 50%] Meshing curve 134 (Line)
Info : [ 50%] Meshing curve 135 (Line)
Info : [ 50%] Meshing curve 136 (Line)
Info : [ 50%] Meshing curve 137 (Line)
Info : [ 50%] Meshing curve 138 (Line)
Info : [ 50%] Meshing curve 139 (Line)
Info : [ 50%] Meshing curve 140 (Line)
Info : [ 50%] Meshing curve 141 (Line)
Info : [ 60%] Meshing curve 142 (Line)
Info : [ 60%] Meshing curve 143 (Line)
Info : [ 60%] Meshing curve 144 (Line)
Info : [ 60%] Meshing curve 145 (Line)
Info : [ 60%] Meshing curve 146 (Line)
Info : [ 60%] Meshing curve 147 (Line)
Info : [ 60%] Meshing curve 148 (Line)
Info : [ 60%] Meshing curve 149 (Line)
Info : [ 70%] Meshing curve 150 (Line)
Info : [ 70%] Meshing curve 151 (Line)
Info : [ 70%] Meshing curve 152 (Line)
Info : [ 70%] Meshing curve 153 (Line)
Info : [ 70%] Meshing curve 154 (Line)
Info : [ 70%] Meshing curve 155 (Line)
Info : [ 70%] Meshing curve 156 (Line)
Info : [ 70%] Meshing curve 157 (Line)
Info : [ 80%] Meshing curve 158 (Line)
Info : [ 80%] Meshing curve 159 (Line)
Info : [ 80%] Meshing curve 160 (Line)
Info : [ 80%] Meshing curve 161 (Line)
Info : [ 80%] Meshing curve 162 (Line)
Info : [ 80%] Meshing curve 163 (Line)
Info : [ 80%] Meshing curve 164 (Line)
Info : [ 80%] Meshing curve 1234 (Line)
Info : [ 90%] Meshing curve 3005 (Circle)
Info : [ 90%] Meshing curve 3006 (Circle)
Info : [ 90%] Meshing curve 3007 (Circle)
Info : [ 90%] Meshing curve 3008 (Circle)
Info : [ 90%] Meshing curve 3015 (Circle)
Info : [ 90%] Meshing curve 3016 (Circle)
Info : [ 90%] Meshing curve 3017 (Circle)
Info : [ 90%] Meshing curve 3018 (Circle)
Info : [100%] Meshing curve 3025 (Circle)
Info : [100%] Meshing curve 3026 (Circle)
Info : [100%] Meshing curve 3027 (Circle)
Info : [100%] Meshing curve 3028 (Circle)
Info : [100%] Meshing curve 3035 (Circle)
Info : [100%] Meshing curve 3036 (Circle)
Info : [100%] Meshing curve 3037 (Circle)
Info : [100%] Meshing curve 3038 (Circle)
Info : Done meshing 1D (Wall 0.00419754s, CPU 0.004118s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 200 (Plane, Delaunay)
Info : [ 0%] Blossom: 2264 internal 88 closed
Info : [ 0%] Blossom recombination completed (Wall 0.0156377s, CPU 0.015536s): 740 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.776443, min Q = 0.460287
Info : [ 30%] Meshing surface 201 (Plane, Delaunay)
Info : [ 30%] Blossom: 2128 internal 208 closed
Info : [ 30%] Blossom recombination completed (Wall 0.0130545s, CPU 0.013s): 744 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.715401, min Q = 0.462407
Info : [ 50%] Meshing surface 202 (Plane, Delaunay)
Info : [ 50%] Blossom: 600 internal 240 closed
Info : [ 50%] Blossom recombination completed (Wall 0.00218844s, CPU 0.002184s): 120 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.983333, min Q = 0.983333
Info : [ 70%] Meshing surface 203 (Plane, Delaunay)
Info : [ 70%] Blossom: 344 internal 248 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00185642s, CPU 0.001883s): 132 quads, 16 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.59063, min Q = 0.274456
Info : [ 90%] Meshing surface 204 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.0645516s, CPU 0.064382s)
Info : 1937 nodes 2682 elements
Info : Writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/GreatCell.msh'
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/domain.vtu (2040 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_Central_sample.vtu (740 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_OuterPart_sample.vtu (1012 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact/physical_group_Rubber_sheath.vtu (288 cells)

%cd {mesh_path}…
(click to toggle)
%cd {mesh_path}
run(
"identifySubdomains -f -m domain.vtu -- "
"physical_group_DSS1.vtu physical_group_DSS1a.vtu "
"physical_group_DSS2.vtu physical_group_DSS2a.vtu "
"physical_group_DSS3.vtu physical_group_DSS3a.vtu "
"physical_group_DSS4.vtu physical_group_DSS4a.vtu "
"physical_group_DSS5.vtu physical_group_DSS5a.vtu "
"physical_group_DSS6.vtu physical_group_DSS6a.vtu "
"physical_group_DSS7.vtu physical_group_DSS7a.vtu "
"physical_group_DSS8.vtu physical_group_DSS8a.vtu "
"physical_group_PEE1.vtu physical_group_PEE1a.vtu "
"physical_group_PEE2.vtu physical_group_PEE2a.vtu "
"physical_group_PEE3.vtu physical_group_PEE3a.vtu "
"physical_group_PEE4.vtu physical_group_PEE4a.vtu "
"physical_group_PEE5.vtu physical_group_PEE5a.vtu "
"physical_group_PEE6.vtu physical_group_PEE6a.vtu "
"physical_group_PEE7.vtu physical_group_PEE7a.vtu "
"physical_group_PEE8.vtu physical_group_PEE8a.vtu "
"physical_group_p_bottom.vtu physical_group_p_left.vtu "
"physical_group_p_right.vtu physical_group_p_top.vtu "
"physical_group_Inlet.vtu ",
shell=True,
check=True,
)
%cd -/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact
info: Mesh reading time: 0.00650056 s
info: MeshNodeSearcher construction time: 8.638e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.467e-06 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000117788 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.9e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.7211e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.9e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.4628e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3796e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.3e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3716e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.51e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3396e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.9e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3085e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.81e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3015e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.61e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.5809e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.11e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3496e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.41e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3336e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.51e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3285e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.1e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.1928e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.5e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3346e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.91e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3506e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.71e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3586e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.91e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3696e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.41e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.4217e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.71e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.4026e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.51e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.4518e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3837e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.11e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.4227e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.6e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3536e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.61e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.634e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.91e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3787e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.41e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3836e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.6e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.7702e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.4117e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.71e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3917e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.71e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3326e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.01e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.641e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.5669e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 2.9e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_bottom' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.0702e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 2.21e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_left' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3676e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.9e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_right' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3185e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 2e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_top' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.5769e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 1.71e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_Inlet' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2855e-05 s
info: identifySubdomains time: 0.00269744 s
info: writing time: 0.0217901 s
info: Entire run time: 0.0312632 s
/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
Run the simulation
Inputs
# Times for load curves…
(click to toggle)
# Times for load curves
times = "0.0 1000. 3500"
simulation_end_time = 3500.0
n_fracture_p_ncs = 0
model_type = "HM1"
output_prefix = "HM1_HM"
# Load
PEE_load_values = {
"A": [10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6, 9.95e6],
"B": [7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6],
"C": [1.0e6, 3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6],
}
for p in materials.values():
p["fluid"][
"injectionFlowRate_Inlet"
] = 2.085e-6 # Injection mass rate kg/s (m_dot = Q_v * rho_f)
prj_file = Path("HM1_HM.prj")
prj = ot.Project(input_file=prj_file, output_file=Path(out_dir, f"{output_prefix}.prj"))Run OGS
sing_ogs_model = SingleOGSModel(…
(click to toggle)
sing_ogs_model = SingleOGSModel(
model=prj,
out_dir=out_dir,
mesh_path=mesh_path,
output_prefix=output_prefix,
method="LIE",
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
materials=materials,
)
# Run simulations
vtu_files_dict = sing_ogs_model.run_simulations_with_fracture(
times=times,
base_project_file=prj_file,
mesh_path=mesh_path,
load_cases=PEE_load_values,
material_names=material_names,
materials=materials,
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
output_prefix=output_prefix,
out_dir=out_dir,
use_b_bar_value="true",
tension_cutoff="true",
)mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM1_HM.prj
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM1_HM_A.prj
============================================================
Running simulation for load case: A with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
[mid=0] Solid density set from material: 2750
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2750
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM1_HM_A.prj
Status: finished successfully.
Execution took 22.634599685668945 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
[mid=0] Solid density set from material: 2650
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2650
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM1_HM_A.prj
Status: finished successfully.
Execution took 23.119516134262085 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM1_HM_B.prj
============================================================
Running simulation for load case: B with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
[mid=0] Solid density set from material: 2750
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2750
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM1_HM_B.prj
Status: finished successfully.
Execution took 38.407310247421265 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
[mid=0] Solid density set from material: 2650
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2650
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM1_HM_B.prj
Status: finished successfully.
Execution took 28.643263339996338 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_intact
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM1_HM_C.prj
============================================================
Running simulation for load case: C with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
[mid=0] Solid density set from material: 2750
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2750
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM1_HM_C.prj
Status: finished successfully.
Execution took 19.51256775856018 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
[mid=0] Solid density set from material: 2650
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2650
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM1_HM_C.prj
Status: finished successfully.
Execution took 22.735936641693115 s
Post-processing
Volumetric strain vs angle at probe circle
plotter = Plotter(…
(click to toggle)
plotter = Plotter(
output_dir=out_dir,
save_extracted_data=True,
)
data_dir = Path("external_data")
external_data = Plotter.load_external_data(data_dir, benchmark_tag="HM1")
plotter.plot_volumetric_strain_vs_angle(
vtu_files_dict,
model_type="HM1",
ylim_range=[-7.5, 2.5],
layout="subplots",
external_data=external_data["strain"],
)

Profiles
Figure below shows the results of this benchmark:
- The trace of effective stress, defined as $\sum_{i=x,y,z} \sigma'_{ii}$,
- and the corresponding volumetric strain plotted versus loading angle for both Greywacke and Gneiss,
These results are shown for the intact rock samples under $\texttt{HM}_1$ loading.
plotter.plot_field_variables(vtu_files_dict)Load: A, File 0, Benchmark: HM1, Material: Gneiss

Load: A, File 1, Benchmark: HM1, Material: Greywacke

Load: B, File 0, Benchmark: HM1, Material: Gneiss

Load: B, File 1, Benchmark: HM1, Material: Greywacke

Load: C, File 0, Benchmark: HM1, Material: Gneiss

Load: C, File 1, Benchmark: HM1, Material: Greywacke

Note: The stress distribution and volumetric strain are shown only in the central zone defined by:
# Plot only inner mesh (within r=0.065 m):
plotter.plot_field_variables(vtu_files_dict, inner=True, r=0.065)Load: A, File 0, Benchmark: HM1, Material: Gneiss

Load: A, File 1, Benchmark: HM1, Material: Greywacke

Load: B, File 0, Benchmark: HM1, Material: Gneiss

Load: B, File 1, Benchmark: HM1, Material: Greywacke

Load: C, File 0, Benchmark: HM1, Material: Gneiss

Load: C, File 1, Benchmark: HM1, Material: Greywacke

Half fractured sample with fluid injection ($\texttt{HM}_{2b}$)
We use all inputs and BCs from $\texttt{M}_{2b}$ for a single wing fracture $\Gamma = [0,\,0.04]\times\{0\}.$ A Dirichlet BC of $p(0.04,0)=3.45\text{ MPa}$ is applied at the fracture’s right tip. After a 3000 s equilibration, fluid is injected at the left tip with $ Q_0^{\mathrm{v}}(0,0)=4.167\times10^{-7}\,\mathrm{m}^3/\mathrm{s}\;(25\text{ ml/min}).$
Mesh Generation
Input
h = 0.005…
(click to toggle)
h = 0.005
meshname = "GreatCell"
mesh_path_embedded = Path(out_dir, "mesh_GreatCell_embeddedFracture").resolve()Boundary meshes
Gmsh (boundary meshes)
msh_file_embedded = mesh_GreatCell_embeddedFracture(…
(click to toggle)
msh_file_embedded = mesh_GreatCell_embeddedFracture(
lc=2 * h,
lc2=h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path_embedded,
meshname=meshname,
mode="BC",
post_process=True,
cmap="viridis",
opacity=0.8,
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 20%] Meshing curve 110 (Line)
Info : [ 20%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 30%] Meshing curve 118 (Line)
Info : [ 30%] Meshing curve 119 (Line)
Info : [ 30%] Meshing curve 120 (Line)
Info : [ 30%] Meshing curve 121 (Line)
Info : [ 30%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 40%] Meshing curve 126 (Line)
Info : [ 40%] Meshing curve 127 (Line)
Info : [ 40%] Meshing curve 128 (Line)
Info : [ 40%] Meshing curve 129 (Line)
Info : [ 40%] Meshing curve 130 (Line)
Info : [ 40%] Meshing curve 131 (Line)
Info : [ 40%] Meshing curve 132 (Line)
Info : [ 40%] Meshing curve 133 (Line)
Info : [ 50%] Meshing curve 134 (Line)
Info : [ 50%] Meshing curve 135 (Line)
Info : [ 50%] Meshing curve 136 (Line)
Info : [ 50%] Meshing curve 137 (Line)
Info : [ 50%] Meshing curve 138 (Line)
Info : [ 50%] Meshing curve 139 (Line)
Info : [ 50%] Meshing curve 140 (Line)
Info : [ 50%] Meshing curve 141 (Line)
Info : [ 60%] Meshing curve 142 (Line)
Info : [ 60%] Meshing curve 143 (Line)
Info : [ 60%] Meshing curve 144 (Line)
Info : [ 60%] Meshing curve 145 (Line)
Info : [ 60%] Meshing curve 146 (Line)
Info : [ 60%] Meshing curve 147 (Line)
Info : [ 60%] Meshing curve 148 (Line)
Info : [ 60%] Meshing curve 149 (Line)
Info : [ 70%] Meshing curve 150 (Line)
Info : [ 70%] Meshing curve 151 (Line)
Info : [ 70%] Meshing curve 152 (Line)
Info : [ 70%] Meshing curve 153 (Line)
Info : [ 70%] Meshing curve 154 (Line)
Info : [ 70%] Meshing curve 155 (Line)
Info : [ 70%] Meshing curve 156 (Line)
Info : [ 70%] Meshing curve 157 (Line)
Info : [ 80%] Meshing curve 158 (Line)
Info : [ 80%] Meshing curve 159 (Line)
Info : [ 80%] Meshing curve 160 (Line)
Info : [ 80%] Meshing curve 161 (Line)
Info : [ 80%] Meshing curve 162 (Line)
Info : [ 80%] Meshing curve 163 (Line)
Info : [ 80%] Meshing curve 164 (Line)
Info : [ 80%] Meshing curve 1234 (Line)
Info : [ 90%] Meshing curve 3005 (Circle)
Info : [ 90%] Meshing curve 3006 (Circle)
Info : [ 90%] Meshing curve 3007 (Circle)
Info : [ 90%] Meshing curve 3008 (Circle)
Info : [ 90%] Meshing curve 3015 (Circle)
Info : [ 90%] Meshing curve 3016 (Circle)
Info : [ 90%] Meshing curve 3017 (Circle)
Info : [ 90%] Meshing curve 3018 (Circle)
Info : [100%] Meshing curve 3025 (Circle)
Info : [100%] Meshing curve 3026 (Circle)
Info : [100%] Meshing curve 3027 (Circle)
Info : [100%] Meshing curve 3028 (Circle)
Info : [100%] Meshing curve 3035 (Circle)
Info : [100%] Meshing curve 3036 (Circle)
Info : [100%] Meshing curve 3037 (Circle)
Info : [100%] Meshing curve 3038 (Circle)
Info : Done meshing 1D (Wall 0.00690915s, CPU 0.004371s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 200 (Plane, Delaunay)
Info : [ 0%] Blossom: 2264 internal 88 closed
Info : [ 0%] Blossom recombination completed (Wall 0.0167279s, CPU 0.016252s): 740 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.776443, min Q = 0.460287
Info : [ 30%] Meshing surface 201 (Plane, Delaunay)
Info : [ 30%] Blossom: 2128 internal 208 closed
Info : [ 30%] Blossom recombination completed (Wall 0.0137989s, CPU 0.012418s): 744 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.715401, min Q = 0.462407
Info : [ 50%] Meshing surface 202 (Plane, Delaunay)
Info : [ 50%] Blossom: 600 internal 240 closed
Info : [ 50%] Blossom recombination completed (Wall 0.00241019s, CPU 0.002337s): 120 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.983333, min Q = 0.983333
Info : [ 70%] Meshing surface 203 (Plane, Delaunay)
Info : [ 70%] Blossom: 344 internal 248 closed
Info : [ 70%] Blossom recombination completed (Wall 0.0020373s, CPU 0.001963s): 132 quads, 16 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.59063, min Q = 0.274456
Info : [ 90%] Meshing surface 204 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.0695233s, CPU 0.066229s)
Info : 1937 nodes 2682 elements
Info : Writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/GreatCell.msh'
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/domain.vtu (96 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_Inlet.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_Outlet_R.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_p_right.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_p_top.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_p_left.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_p_bottom.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE1.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE2.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE3.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE4.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE5.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE6.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE7.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE8.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE1a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE2a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE3a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE4a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE5a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE6a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE7a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_PEE8a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS1.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS2.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS3.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS4.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS5.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS6.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS7.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS8.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS1a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS2a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS3a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS4a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS5a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS6a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS7a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_DSS8a.vtu (2 cells)

Computational domain mesh
Gmsh
msh_file_volume_embedded = mesh_GreatCell_embeddedFracture(…
(click to toggle)
msh_file_volume_embedded = mesh_GreatCell_embeddedFracture(
lc=2 * h,
lc2=h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path_embedded,
meshname=meshname,
mode="domain",
post_process=True,
cmap="viridis",
opacity=0.8,
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 20%] Meshing curve 110 (Line)
Info : [ 20%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 30%] Meshing curve 118 (Line)
Info : [ 30%] Meshing curve 119 (Line)
Info : [ 30%] Meshing curve 120 (Line)
Info : [ 30%] Meshing curve 121 (Line)
Info : [ 30%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 40%] Meshing curve 126 (Line)
Info : [ 40%] Meshing curve 127 (Line)
Info : [ 40%] Meshing curve 128 (Line)
Info : [ 40%] Meshing curve 129 (Line)
Info : [ 40%] Meshing curve 130 (Line)
Info : [ 40%] Meshing curve 131 (Line)
Info : [ 40%] Meshing curve 132 (Line)
Info : [ 40%] Meshing curve 133 (Line)
Info : [ 50%] Meshing curve 134 (Line)
Info : [ 50%] Meshing curve 135 (Line)
Info : [ 50%] Meshing curve 136 (Line)
Info : [ 50%] Meshing curve 137 (Line)
Info : [ 50%] Meshing curve 138 (Line)
Info : [ 50%] Meshing curve 139 (Line)
Info : [ 50%] Meshing curve 140 (Line)
Info : [ 50%] Meshing curve 141 (Line)
Info : [ 60%] Meshing curve 142 (Line)
Info : [ 60%] Meshing curve 143 (Line)
Info : [ 60%] Meshing curve 144 (Line)
Info : [ 60%] Meshing curve 145 (Line)
Info : [ 60%] Meshing curve 146 (Line)
Info : [ 60%] Meshing curve 147 (Line)
Info : [ 60%] Meshing curve 148 (Line)
Info : [ 60%] Meshing curve 149 (Line)
Info : [ 70%] Meshing curve 150 (Line)
Info : [ 70%] Meshing curve 151 (Line)
Info : [ 70%] Meshing curve 152 (Line)
Info : [ 70%] Meshing curve 153 (Line)
Info : [ 70%] Meshing curve 154 (Line)
Info : [ 70%] Meshing curve 155 (Line)
Info : [ 70%] Meshing curve 156 (Line)
Info : [ 70%] Meshing curve 157 (Line)
Info : [ 80%] Meshing curve 158 (Line)
Info : [ 80%] Meshing curve 159 (Line)
Info : [ 80%] Meshing curve 160 (Line)
Info : [ 80%] Meshing curve 161 (Line)
Info : [ 80%] Meshing curve 162 (Line)
Info : [ 80%] Meshing curve 163 (Line)
Info : [ 80%] Meshing curve 164 (Line)
Info : [ 80%] Meshing curve 1234 (Line)
Info : [ 90%] Meshing curve 3005 (Circle)
Info : [ 90%] Meshing curve 3006 (Circle)
Info : [ 90%] Meshing curve 3007 (Circle)
Info : [ 90%] Meshing curve 3008 (Circle)
Info : [ 90%] Meshing curve 3015 (Circle)
Info : [ 90%] Meshing curve 3016 (Circle)
Info : [ 90%] Meshing curve 3017 (Circle)
Info : [ 90%] Meshing curve 3018 (Circle)
Info : [100%] Meshing curve 3025 (Circle)
Info : [100%] Meshing curve 3026 (Circle)
Info : [100%] Meshing curve 3027 (Circle)
Info : [100%] Meshing curve 3028 (Circle)
Info : [100%] Meshing curve 3035 (Circle)
Info : [100%] Meshing curve 3036 (Circle)
Info : [100%] Meshing curve 3037 (Circle)
Info : [100%] Meshing curve 3038 (Circle)
Info : Done meshing 1D (Wall 0.00478795s, CPU 0.004543s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 200 (Plane, Delaunay)
Info : [ 0%] Blossom: 2264 internal 88 closed
Info : [ 0%] Blossom recombination completed (Wall 0.0157308s, CPU 0.015656s): 740 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.776443, min Q = 0.460287
Info : [ 30%] Meshing surface 201 (Plane, Delaunay)
Info : [ 30%] Blossom: 2128 internal 208 closed
Info : [ 30%] Blossom recombination completed (Wall 0.0133307s, CPU 0.013264s): 744 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.715401, min Q = 0.462407
Info : [ 50%] Meshing surface 202 (Plane, Delaunay)
Info : [ 50%] Blossom: 600 internal 240 closed
Info : [ 50%] Blossom recombination completed (Wall 0.0023569s, CPU 0.002332s): 120 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.983333, min Q = 0.983333
Info : [ 70%] Meshing surface 203 (Plane, Delaunay)
Info : [ 70%] Blossom: 344 internal 248 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00198808s, CPU 0.001969s): 132 quads, 16 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.59063, min Q = 0.274456
Info : [ 90%] Meshing surface 204 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.0663461s, CPU 0.065807s)
Info : 1937 nodes 2682 elements
Info : Writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/GreatCell.msh'
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/domain.vtu (2048 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_Fracture.vtu (8 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_Central_sample.vtu (740 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_OuterPart_sample.vtu (1012 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture/physical_group_Rubber_sheath.vtu (288 cells)

%cd {mesh_path_embedded}…
(click to toggle)
%cd {mesh_path_embedded}
!pwd
run(
"identifySubdomains -f -m domain.vtu -- "
"physical_group_DSS1.vtu physical_group_DSS1a.vtu "
"physical_group_DSS2.vtu physical_group_DSS2a.vtu "
"physical_group_DSS3.vtu physical_group_DSS3a.vtu "
"physical_group_DSS4.vtu physical_group_DSS4a.vtu "
"physical_group_DSS5.vtu physical_group_DSS5a.vtu "
"physical_group_DSS6.vtu physical_group_DSS6a.vtu "
"physical_group_DSS7.vtu physical_group_DSS7a.vtu "
"physical_group_DSS8.vtu physical_group_DSS8a.vtu "
"physical_group_PEE1.vtu physical_group_PEE1a.vtu "
"physical_group_PEE2.vtu physical_group_PEE2a.vtu "
"physical_group_PEE3.vtu physical_group_PEE3a.vtu "
"physical_group_PEE4.vtu physical_group_PEE4a.vtu "
"physical_group_PEE5.vtu physical_group_PEE5a.vtu "
"physical_group_PEE6.vtu physical_group_PEE6a.vtu "
"physical_group_PEE7.vtu physical_group_PEE7a.vtu "
"physical_group_PEE8.vtu physical_group_PEE8a.vtu "
"physical_group_p_bottom.vtu physical_group_p_left.vtu "
"physical_group_p_right.vtu physical_group_p_top.vtu "
"physical_group_Inlet.vtu physical_group_Outlet_R.vtu ",
shell=True,
check=True,
)
%cd -/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture
/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture
info: Mesh reading time: 0.0069065 s
info: MeshNodeSearcher construction time: 8.4588e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.738e-06 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000120702 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.81e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.8633e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.21e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.4066e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.6e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2825e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.4e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2445e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.81e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.4457e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.71e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2094e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.71e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.645e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.81e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.1963e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.8e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2193e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.4e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.1964e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.31e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.8884e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.71e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2304e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.9e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2565e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.31e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2364e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.5e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2173e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.3465e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2734e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.11e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2805e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.11e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2574e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.3e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.4998e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.11e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.598e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.71e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2495e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.01e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2274e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.11e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2304e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.71e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.2434e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.21e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000186521 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.5038e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.0291e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.21e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.4307e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.31e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.0401e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.61e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.6801e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 2.9e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_bottom' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.615e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.41e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_left' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.4558e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 2.8e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_right' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.7482e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 2.9e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_top' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.625e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.01e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_Inlet' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.4598e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.61e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_Outlet_R' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 6.5619e-05 s
info: identifySubdomains time: 0.00372939 s
info: writing time: 0.0240847 s
info: Entire run time: 0.0356099 s
/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
Run the simulation
Input
# Times for load curves…
(click to toggle)
# Times for load curves
times = "0.0 1000. 3500"
simulation_end_time = 3500.0
n_fracture_p_ncs = 3
model_type = "HM2b"
output_prefix = "HM2b_LIE"
# Load
PEE_load_values = {
"A": [10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6, 9.95e6],
"E": [9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6],
"B": [7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6],
"F": [3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6],
"C": [1.0e6, 3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6],
}
for p in materials.values():
p["fluid"][
"injectionFlowRate_Inlet"
] = 4.167e-7 # Injection flow rate m³/s (125 ml/min)
# Project file
prj_file = Path("HM2b_LIE.prj")
prj = ot.Project(input_file=prj_file, output_file=Path(out_dir, f"{output_prefix}.prj"))Run OGS
sing_ogs_model = SingleOGSModel(…
(click to toggle)
sing_ogs_model = SingleOGSModel(
model=prj,
out_dir=out_dir,
mesh_path=mesh_path_embedded,
output_prefix=output_prefix,
method="LIE",
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
materials=materials,
)
# Run simulations
vtu_files_dict_embedded = sing_ogs_model.run_simulations_with_fracture(
times=times,
base_project_file=prj_file,
mesh_path=mesh_path_embedded,
load_cases=PEE_load_values,
material_names=material_names,
materials=materials,
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
output_prefix=output_prefix,
out_dir=out_dir,
use_b_bar_value="true",
tension_cutoff="true",
)mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE.prj
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_A.prj
============================================================
Running simulation for load case: A with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
[mid=0] Solid density set from material: 2750
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2750
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2750
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_A.prj
Status: finished successfully.
Execution took 34.44262957572937 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
[mid=0] Solid density set from material: 2650
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2650
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2650
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_A.prj
Status: finished successfully.
Execution took 38.48575305938721 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_E.prj
============================================================
Running simulation for load case: E with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
[mid=0] Solid density set from material: 2750
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2750
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2750
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_E.prj
Status: finished successfully.
Execution took 26.490394353866577 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
[mid=0] Solid density set from material: 2650
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2650
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2650
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_E.prj
Status: finished successfully.
Execution took 31.612836360931396 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_B.prj
============================================================
Running simulation for load case: B with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
[mid=0] Solid density set from material: 2750
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2750
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2750
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_B.prj
Status: finished successfully.
Execution took 27.362358331680298 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
[mid=0] Solid density set from material: 2650
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2650
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2650
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_B.prj
Status: finished successfully.
Execution took 48.364235639572144 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_F.prj
============================================================
Running simulation for load case: F with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
[mid=0] Solid density set from material: 2750
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2750
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2750
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_F.prj
Status: finished successfully.
Execution took 30.788437128067017 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
[mid=0] Solid density set from material: 2650
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2650
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2650
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_F.prj
Status: finished successfully.
Execution took 52.581350803375244 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_embeddedFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_C.prj
============================================================
Running simulation for load case: C with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
[mid=0] Solid density set from material: 2750
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2750
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2750
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_C.prj
Status: finished successfully.
Execution took 73.99682092666626 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
[mid=0] Solid density set from material: 2650
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2650
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2650
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2b_LIE_C.prj
Status: finished successfully.
Execution took 61.26739239692688 s
Post-processing
data_dir = Path("external_data")…
(click to toggle)
data_dir = Path("external_data")
external_data = Plotter.load_external_data(data_dir, benchmark_tag="HM2b")
custom_cb = {
"u": {"vmin": 0, "vmax": 0.25, "cmap": "Greens"},
"stress": {"vmin": -20, "vmax": 10, "cmap": "coolwarm"},
"strain": {"vmin": -0.05, "vmax": 0, "cmap": "RdBu_r"},
"pressure": {"vmin": 0.1, "vmax": 8, "cmap": "jet"},
}
plotter = Plotter(
output_dir=out_dir,
colorbar_opts=custom_cb,
save_extracted_data=True,
)
plotter.plot_volumetric_strain_vs_angle(
vtu_files_dict_embedded,
model_type="HM2b",
ylim_range=[-7.5, 2.5],
layout="subplots", # "single" or "subplots"
external_data=external_data["strain"],
)

plotter.plot_field_variables(vtu_files_dict_embedded)Load: A, File 0, Benchmark: HM2b, Material: Gneiss

Load: A, File 1, Benchmark: HM2b, Material: Greywacke

Load: E, File 0, Benchmark: HM2b, Material: Gneiss

Load: E, File 1, Benchmark: HM2b, Material: Greywacke

Load: B, File 0, Benchmark: HM2b, Material: Gneiss

Load: B, File 1, Benchmark: HM2b, Material: Greywacke

Load: F, File 0, Benchmark: HM2b, Material: Gneiss

Load: F, File 1, Benchmark: HM2b, Material: Greywacke

Load: C, File 0, Benchmark: HM2b, Material: Gneiss

Load: C, File 1, Benchmark: HM2b, Material: Greywacke

plotter.material_names = ["Gneiss", "Greywacke"]…
(click to toggle)
plotter.material_names = ["Gneiss", "Greywacke"]
plotter.vtu_file_names = {"LIE": vtu_files_dict_embedded}
plotter.plot_avg_width_vs_stress(
benchmark_tag="HM2b",
metric="width",
methods_to_include=["LIE"],
pee_load_values=PEE_load_values,
external_data=external_data["average"],
)
plotter.plot_avg_width_vs_stress(
benchmark_tag="HM2b",
metric="permeability",
methods_to_include=["LIE"],
pee_load_values=PEE_load_values,
external_data=external_data["average"],
)


array([ 1000000. , 2318019.48466054, 5500000. ,
8681980.51533946, 10000000. ])
lie_profiles = {}…
(click to toggle)
lie_profiles = {}
for load_case, file_list in plotter.vtu_file_names["LIE"].items():
lie_profiles[load_case] = plotter.extract_lie_aperture_from_list(
file_list,
plotter.material_names,
)
plotter.plot_fracture_aperture_profiles(
widthProfile=lie_profiles,
benchmark_tag="HM2b",
downsample=1,
ylim=(0, 10e-6),
method_label="LIE",
external_data=external_data["widthProfile"],
)





Fully fractured sample with fluid injection ($\texttt{HM}_{2a}$)
We performed five hydro-mechanical (HMca) simulations with different angles between the second principal stress direction (σ₂ = 10 MPa) and the PEE 5 fracture plane $\Gamma = [-0.094,\,0.094]\times\{0\}.$ The initial pore pressure was uniform at $p_0 = 0.1$ MPa, with Dirichlet conditions $p(-0.094,0)=p(0.094,0)=3.45\text{ MPa}.$ After equilibrating for 3000 s, we injected fluid at the center at a rate $ Q_0^{\text{v}}(0,0)=4.167\times10^{-7}\,\mathrm{m}^3/\mathrm{s}\;(25\,\mathrm{ml/min})$ for 500 s, starting from an initial fracture aperture $w_{\text{ini}}=1\times10^{-6}$ m.
Mesh Generation
Input
h = 0.005…
(click to toggle)
h = 0.005
meshname = "GreatCell"
mesh_path_full = Path(".", out_dir, "mesh_GreatCell_fullFracture").resolve()Boundary meshes
Gmsh
msh_file_full = mesh_GreatCell_fullFracture(…
(click to toggle)
msh_file_full = mesh_GreatCell_fullFracture(
lc=2 * h,
lc2=h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path_full,
meshname=meshname,
mode="BC",
post_process=True,
cmap="viridis",
opacity=0.8,
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 20%] Meshing curve 110 (Line)
Info : [ 20%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 20%] Meshing curve 118 (Line)
Info : [ 30%] Meshing curve 119 (Line)
Info : [ 30%] Meshing curve 120 (Line)
Info : [ 30%] Meshing curve 121 (Line)
Info : [ 30%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 30%] Meshing curve 126 (Line)
Info : [ 30%] Meshing curve 127 (Line)
Info : [ 40%] Meshing curve 128 (Line)
Info : [ 40%] Meshing curve 129 (Line)
Info : [ 40%] Meshing curve 130 (Line)
Info : [ 40%] Meshing curve 131 (Line)
Info : [ 40%] Meshing curve 132 (Line)
Info : [ 40%] Meshing curve 133 (Line)
Info : [ 40%] Meshing curve 134 (Line)
Info : [ 40%] Meshing curve 135 (Line)
Info : [ 40%] Meshing curve 136 (Line)
Info : [ 50%] Meshing curve 137 (Line)
Info : [ 50%] Meshing curve 138 (Line)
Info : [ 50%] Meshing curve 139 (Line)
Info : [ 50%] Meshing curve 140 (Line)
Info : [ 50%] Meshing curve 141 (Line)
Info : [ 50%] Meshing curve 142 (Line)
Info : [ 50%] Meshing curve 143 (Line)
Info : [ 50%] Meshing curve 144 (Line)
Info : [ 60%] Meshing curve 145 (Line)
Info : [ 60%] Meshing curve 146 (Line)
Info : [ 60%] Meshing curve 147 (Line)
Info : [ 60%] Meshing curve 148 (Line)
Info : [ 60%] Meshing curve 149 (Line)
Info : [ 60%] Meshing curve 150 (Line)
Info : [ 60%] Meshing curve 151 (Line)
Info : [ 60%] Meshing curve 152 (Line)
Info : [ 60%] Meshing curve 153 (Line)
Info : [ 70%] Meshing curve 154 (Line)
Info : [ 70%] Meshing curve 155 (Line)
Info : [ 70%] Meshing curve 156 (Line)
Info : [ 70%] Meshing curve 157 (Line)
Info : [ 70%] Meshing curve 158 (Line)
Info : [ 70%] Meshing curve 159 (Line)
Info : [ 70%] Meshing curve 160 (Line)
Info : [ 70%] Meshing curve 161 (Line)
Info : [ 70%] Meshing curve 162 (Line)
Info : [ 80%] Meshing curve 163 (Line)
Info : [ 80%] Meshing curve 164 (Line)
Info : [ 80%] Meshing curve 3005 (Circle)
Info : [ 80%] Meshing curve 3006 (Circle)
Info : [ 80%] Meshing curve 3007 (Circle)
Info : [ 80%] Meshing curve 3008 (Circle)
Info : [ 80%] Meshing curve 3015 (Circle)
Info : [ 80%] Meshing curve 3016 (Circle)
Info : [ 80%] Meshing curve 3017 (Circle)
Info : [ 90%] Meshing curve 3018 (Circle)
Info : [ 90%] Meshing curve 3025 (Circle)
Info : [ 90%] Meshing curve 3026 (Circle)
Info : [ 90%] Meshing curve 3027 (Circle)
Info : [ 90%] Meshing curve 3028 (Circle)
Info : [ 90%] Meshing curve 3035 (Circle)
Info : [ 90%] Meshing curve 3036 (Circle)
Info : [ 90%] Meshing curve 3037 (Circle)
Info : [ 90%] Meshing curve 3038 (Circle)
Info : [100%] Meshing curve 8000 (Line)
Info : [100%] Meshing curve 8001 (Line)
Info : [100%] Meshing curve 8002 (Line)
Info : [100%] Meshing curve 8003 (Line)
Info : [100%] Meshing curve 8004 (Line)
Info : [100%] Meshing curve 8005 (Line)
Info : [100%] Meshing curve 8006 (Line)
Info : [100%] Meshing curve 8007 (Line)
Info : Done meshing 1D (Wall 0.0055233s, CPU 0.004325s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 200 (Plane, Delaunay)
Info : [ 0%] Blossom: 1203 internal 72 closed
Info : [ 0%] Blossom recombination completed (Wall 0.00949125s, CPU 0.009165s): 400 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.743465, min Q = 0.472764
Info : [ 20%] Meshing surface 201 (Plane, Delaunay)
Info : [ 20%] Blossom: 1070 internal 116 closed
Info : [ 20%] Blossom recombination completed (Wall 0.00783522s, CPU 0.007793s): 370 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.751278, min Q = 0.477496
Info : [ 30%] Meshing surface 202 (Plane, Delaunay)
Info : [ 30%] Blossom: 301 internal 124 closed
Info : [ 30%] Blossom recombination completed (Wall 0.00241503s, CPU 0.002395s): 92 quads, 58 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.475993, min Q = 0.301098
Info : [ 40%] Meshing surface 203 (Plane, Delaunay)
Info : [ 40%] Blossom: 171 internal 126 closed
Info : [ 40%] Blossom recombination completed (Wall 0.00103092s, CPU 0.001018s): 66 quads, 8 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.59063, min Q = 0.274456
Info : [ 50%] Meshing surface 204 (Plane, Delaunay)
Info : [ 50%] Blossom: 1203 internal 72 closed
Info : [ 50%] Blossom recombination completed (Wall 0.00926757s, CPU 0.009218s): 400 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.743465, min Q = 0.472764
Info : [ 60%] Meshing surface 205 (Plane, Delaunay)
Info : [ 60%] Blossom: 1070 internal 116 closed
Info : [ 60%] Blossom recombination completed (Wall 0.0076579s, CPU 0.007613s): 370 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.751278, min Q = 0.477496
Info : [ 70%] Meshing surface 206 (Plane, Delaunay)
Info : [ 70%] Blossom: 301 internal 124 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00241012s, CPU 0.002389s): 92 quads, 58 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.475993, min Q = 0.301098
Info : [ 80%] Meshing surface 207 (Plane, Delaunay)
Info : [ 80%] Blossom: 171 internal 126 closed
Info : [ 80%] Blossom recombination completed (Wall 0.00106142s, CPU 0.001046s): 66 quads, 8 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.59063, min Q = 0.274456
Info : [ 90%] Meshing surface 208 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.0763732s, CPU 0.074543s)
Info : 2115 nodes 2955 elements
Info : Writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/GreatCell.msh'
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/domain.vtu (96 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_Inlet.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_p_right.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_p_top.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_p_left.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_p_bottom.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_Outlet_R.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_Outlet_L.vtu (1 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE1.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE2.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE3.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE4.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE5.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE6.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE7.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE8.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE1a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE2a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE3a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE4a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE5a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE6a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE7a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_PEE8a.vtu (4 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS1.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS2.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS3.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS4.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS5.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS6.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS7.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS8.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS1a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS2a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS3a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS4a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS5a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS6a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS7a.vtu (2 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_DSS8a.vtu (2 cells)

Computational domain mesh
Gmsh
msh_file_full = mesh_GreatCell_fullFracture(…
(click to toggle)
msh_file_full = mesh_GreatCell_fullFracture(
lc=2 * h,
lc2=h,
r0=0.097,
r1=0.094,
r2=0.090,
r3=0.065,
out_dir=mesh_path_full,
meshname=meshname,
mode="domain",
post_process=True,
cmap="viridis",
opacity=0.8,
)Info : Meshing 1D...
Info : [ 0%] Meshing curve 101 (Line)
Info : [ 10%] Meshing curve 102 (Line)
Info : [ 10%] Meshing curve 103 (Line)
Info : [ 10%] Meshing curve 104 (Line)
Info : [ 10%] Meshing curve 105 (Line)
Info : [ 10%] Meshing curve 106 (Line)
Info : [ 10%] Meshing curve 107 (Line)
Info : [ 10%] Meshing curve 108 (Line)
Info : [ 10%] Meshing curve 109 (Line)
Info : [ 20%] Meshing curve 110 (Line)
Info : [ 20%] Meshing curve 111 (Line)
Info : [ 20%] Meshing curve 112 (Line)
Info : [ 20%] Meshing curve 113 (Line)
Info : [ 20%] Meshing curve 114 (Line)
Info : [ 20%] Meshing curve 115 (Line)
Info : [ 20%] Meshing curve 116 (Line)
Info : [ 20%] Meshing curve 117 (Line)
Info : [ 20%] Meshing curve 118 (Line)
Info : [ 30%] Meshing curve 119 (Line)
Info : [ 30%] Meshing curve 120 (Line)
Info : [ 30%] Meshing curve 121 (Line)
Info : [ 30%] Meshing curve 122 (Line)
Info : [ 30%] Meshing curve 123 (Line)
Info : [ 30%] Meshing curve 124 (Line)
Info : [ 30%] Meshing curve 125 (Line)
Info : [ 30%] Meshing curve 126 (Line)
Info : [ 30%] Meshing curve 127 (Line)
Info : [ 40%] Meshing curve 128 (Line)
Info : [ 40%] Meshing curve 129 (Line)
Info : [ 40%] Meshing curve 130 (Line)
Info : [ 40%] Meshing curve 131 (Line)
Info : [ 40%] Meshing curve 132 (Line)
Info : [ 40%] Meshing curve 133 (Line)
Info : [ 40%] Meshing curve 134 (Line)
Info : [ 40%] Meshing curve 135 (Line)
Info : [ 40%] Meshing curve 136 (Line)
Info : [ 50%] Meshing curve 137 (Line)
Info : [ 50%] Meshing curve 138 (Line)
Info : [ 50%] Meshing curve 139 (Line)
Info : [ 50%] Meshing curve 140 (Line)
Info : [ 50%] Meshing curve 141 (Line)
Info : [ 50%] Meshing curve 142 (Line)
Info : [ 50%] Meshing curve 143 (Line)
Info : [ 50%] Meshing curve 144 (Line)
Info : [ 60%] Meshing curve 145 (Line)
Info : [ 60%] Meshing curve 146 (Line)
Info : [ 60%] Meshing curve 147 (Line)
Info : [ 60%] Meshing curve 148 (Line)
Info : [ 60%] Meshing curve 149 (Line)
Info : [ 60%] Meshing curve 150 (Line)
Info : [ 60%] Meshing curve 151 (Line)
Info : [ 60%] Meshing curve 152 (Line)
Info : [ 60%] Meshing curve 153 (Line)
Info : [ 70%] Meshing curve 154 (Line)
Info : [ 70%] Meshing curve 155 (Line)
Info : [ 70%] Meshing curve 156 (Line)
Info : [ 70%] Meshing curve 157 (Line)
Info : [ 70%] Meshing curve 158 (Line)
Info : [ 70%] Meshing curve 159 (Line)
Info : [ 70%] Meshing curve 160 (Line)
Info : [ 70%] Meshing curve 161 (Line)
Info : [ 70%] Meshing curve 162 (Line)
Info : [ 80%] Meshing curve 163 (Line)
Info : [ 80%] Meshing curve 164 (Line)
Info : [ 80%] Meshing curve 3005 (Circle)
Info : [ 80%] Meshing curve 3006 (Circle)
Info : [ 80%] Meshing curve 3007 (Circle)
Info : [ 80%] Meshing curve 3008 (Circle)
Info : [ 80%] Meshing curve 3015 (Circle)
Info : [ 80%] Meshing curve 3016 (Circle)
Info : [ 80%] Meshing curve 3017 (Circle)
Info : [ 90%] Meshing curve 3018 (Circle)
Info : [ 90%] Meshing curve 3025 (Circle)
Info : [ 90%] Meshing curve 3026 (Circle)
Info : [ 90%] Meshing curve 3027 (Circle)
Info : [ 90%] Meshing curve 3028 (Circle)
Info : [ 90%] Meshing curve 3035 (Circle)
Info : [ 90%] Meshing curve 3036 (Circle)
Info : [ 90%] Meshing curve 3037 (Circle)
Info : [ 90%] Meshing curve 3038 (Circle)
Info : [100%] Meshing curve 8000 (Line)
Info : [100%] Meshing curve 8001 (Line)
Info : [100%] Meshing curve 8002 (Line)
Info : [100%] Meshing curve 8003 (Line)
Info : [100%] Meshing curve 8004 (Line)
Info : [100%] Meshing curve 8005 (Line)
Info : [100%] Meshing curve 8006 (Line)
Info : [100%] Meshing curve 8007 (Line)
Info : Done meshing 1D (Wall 0.00600189s, CPU 0.005503s)
Info : Meshing 2D...
Info : [ 0%] Meshing surface 200 (Plane, Delaunay)
Info : [ 0%] Blossom: 1203 internal 72 closed
Info : [ 0%] Blossom recombination completed (Wall 0.00918487s, CPU 0.009124s): 400 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.743465, min Q = 0.472764
Info : [ 20%] Meshing surface 201 (Plane, Delaunay)
Info : [ 20%] Blossom: 1070 internal 116 closed
Info : [ 20%] Blossom recombination completed (Wall 0.0075743s, CPU 0.007528s): 370 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.751278, min Q = 0.477496
Info : [ 30%] Meshing surface 202 (Plane, Delaunay)
Info : [ 30%] Blossom: 301 internal 124 closed
Info : [ 30%] Blossom recombination completed (Wall 0.00233021s, CPU 0.002303s): 92 quads, 58 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.475993, min Q = 0.301098
Info : [ 40%] Meshing surface 203 (Plane, Delaunay)
Info : [ 40%] Blossom: 171 internal 126 closed
Info : [ 40%] Blossom recombination completed (Wall 0.000974432s, CPU 0.000957s): 66 quads, 8 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.59063, min Q = 0.274456
Info : [ 50%] Meshing surface 204 (Plane, Delaunay)
Info : [ 50%] Blossom: 1203 internal 72 closed
Info : [ 50%] Blossom recombination completed (Wall 0.00912957s, CPU 0.009065s): 400 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.743465, min Q = 0.472764
Info : [ 60%] Meshing surface 205 (Plane, Delaunay)
Info : [ 60%] Blossom: 1070 internal 116 closed
Info : [ 60%] Blossom recombination completed (Wall 0.00773701s, CPU 0.007675s): 370 quads, 0 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.751278, min Q = 0.477496
Info : [ 70%] Meshing surface 206 (Plane, Delaunay)
Info : [ 70%] Blossom: 301 internal 124 closed
Info : [ 70%] Blossom recombination completed (Wall 0.00236588s, CPU 0.002343s): 92 quads, 58 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.475993, min Q = 0.301098
Info : [ 80%] Meshing surface 207 (Plane, Delaunay)
Info : [ 80%] Blossom: 171 internal 126 closed
Info : [ 80%] Blossom recombination completed (Wall 0.00101117s, CPU 0.000994s): 66 quads, 8 triangles, 0 invalid quads, 0 quads with Q < 0.1, avg Q = 0.59063, min Q = 0.274456
Info : [ 90%] Meshing surface 208 (Plane, Delaunay)
Info : Done meshing 2D (Wall 0.0745847s, CPU 0.073937s)
Info : 2115 nodes 2955 elements
Info : Writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/GreatCell.msh'...
Info : Done writing '/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/GreatCell.msh'
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/domain.vtu (2320 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_fracture.vtu (44 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_Central_sample.vtu (800 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_OuterPart_sample.vtu (1188 cells)
Saved: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture/physical_group_Rubber_sheath.vtu (288 cells)

%cd {mesh_path_full}…
(click to toggle)
%cd {mesh_path_full}
run(
"identifySubdomains -f -m domain.vtu -- "
"physical_group_DSS1.vtu physical_group_DSS1a.vtu "
"physical_group_DSS2.vtu physical_group_DSS2a.vtu "
"physical_group_DSS3.vtu physical_group_DSS3a.vtu "
"physical_group_DSS4.vtu physical_group_DSS4a.vtu "
"physical_group_DSS5.vtu physical_group_DSS5a.vtu "
"physical_group_DSS6.vtu physical_group_DSS6a.vtu "
"physical_group_DSS7.vtu physical_group_DSS7a.vtu "
"physical_group_DSS8.vtu physical_group_DSS8a.vtu "
"physical_group_PEE1.vtu physical_group_PEE1a.vtu "
"physical_group_PEE2.vtu physical_group_PEE2a.vtu "
"physical_group_PEE3.vtu physical_group_PEE3a.vtu "
"physical_group_PEE4.vtu physical_group_PEE4a.vtu "
"physical_group_PEE5.vtu physical_group_PEE5a.vtu "
"physical_group_PEE6.vtu physical_group_PEE6a.vtu "
"physical_group_PEE7.vtu physical_group_PEE7a.vtu "
"physical_group_PEE8.vtu physical_group_PEE8a.vtu "
"physical_group_p_bottom.vtu physical_group_p_left.vtu "
"physical_group_p_right.vtu physical_group_p_top.vtu "
"physical_group_Inlet.vtu physical_group_Outlet_R.vtu "
"physical_group_Outlet_L.vtu ",
shell=True,
check=True,
)
%cd -/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture
info: Mesh reading time: 0.00716258 s
info: MeshNodeSearcher construction time: 8.8964e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.008e-06 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 0.000138259 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.01e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS1a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.893e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.31e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.6486e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.51e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS2a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.293e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.2e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.3341e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.71e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS3a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.0417e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.11e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.7557e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.91e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS4a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.3611e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.21e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.0707e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.41e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS5a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.27e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.8e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.0226e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.61e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS6a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.3952e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.21e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.1699e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.41e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS7a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.1007e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.8e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.1478e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.81e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_DSS8a' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.1277e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.51e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.3521e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 8.21e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE1a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.2079e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.01e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.1518e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.51e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE2a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.9109e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.71e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.254e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 9.51e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE3a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.282e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.31e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.5514e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.31e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE4a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.1688e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.21e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.1719e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE5a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.1948e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.61e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.1989e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.21e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE6a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.2529e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 6.01e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.2129e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.21e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE7a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.25e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.81e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.2479e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 5.5e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_PEE8a' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.7016e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_bottom' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.304e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.7e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_left' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.5073e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 2.7e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_right' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 8.3566e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.7e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_p_top' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.4142e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.81e-07 s
info: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_Inlet' and it is equal to the newly computed values.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.4353e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 4.11e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_Outlet_R' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.3311e-05 s
info: identifySubdomainMesh(): identifySubdomainMeshNodes took 3.1e-07 s
warning: There is already a 'bulk_node_ids' property present in the subdomain mesh 'physical_group_Outlet_L' and it is not equal to the newly computed values.
info: Overwriting 'bulk_node_ids' property.
info: identifySubdomainMesh(): identifySubdomainMeshElements took 7.5504e-05 s
info: identifySubdomains time: 0.00334499 s
info: writing time: 0.0247409 s
info: Entire run time: 0.0357519 s
/var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
Run the simulation
Input
# Times for load curves…
(click to toggle)
# Times for load curves
times = "0.0 1000. 3500"
simulation_end_time = 3500.0
n_fracture_p_ncs = 3
model_type = "HM2a"
output_prefix = "HM2a_LIE"
# Load
PEE_load_values = {
"A": [10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6, 9.95e6],
"E": [9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6, 7.80e6],
"B": [7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6, 3.82e6],
"F": [3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6, 1.0e6],
"C": [1.0e6, 3.82e6, 7.80e6, 9.95e6, 10.0e6, 6.64e6, 4.46e6, 1.17e6],
}
for p in materials.values():
p["fluid"][
"injectionFlowRate_Inlet"
] = 4.167e-7 # Injection flow rate m³/s (125 ml/min)
# Project file
prj_file = Path("HM2a_LIE.prj")
prj = ot.Project(input_file=prj_file, output_file=Path(out_dir, f"{output_prefix}.prj"))Run OGS
sing_ogs_model = SingleOGSModel(
model=prj,
out_dir=out_dir,
mesh_path=mesh_path_full,
output_prefix=output_prefix,
method="LIE",
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
materials=materials,
)
# Run simulations
vtu_files_dict_full = sing_ogs_model.run_simulations_with_fracture(
times=times,
base_project_file=prj_file,
mesh_path=mesh_path_full,
load_cases=PEE_load_values,
material_names=material_names,
materials=materials,
n_fracture_p_ncs=n_fracture_p_ncs,
model_type=model_type,
output_prefix=output_prefix,
out_dir=out_dir,
use_b_bar_value="true",
tension_cutoff="true",
)mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE.prj
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_A.prj
============================================================
Running simulation for load case: A with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
[mid=0] Solid density set from material: 2750
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2750
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2750
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_A.prj
Status: finished successfully.
Execution took 71.8872344493866 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
[mid=0] Solid density set from material: 2650
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2650
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2650
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_A.prj
Status: finished successfully.
Execution took 106.43588924407959 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_E.prj
============================================================
Running simulation for load case: E with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
[mid=0] Solid density set from material: 2750
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2750
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2750
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_E.prj
Status: finished successfully.
Execution took 80.15128135681152 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
[mid=0] Solid density set from material: 2650
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2650
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2650
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_E.prj
Status: finished successfully.
Execution took 79.60322976112366 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_B.prj
============================================================
Running simulation for load case: B with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
[mid=0] Solid density set from material: 2750
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2750
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2750
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_B.prj
Status: finished successfully.
Execution took 70.14914035797119 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
[mid=0] Solid density set from material: 2650
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2650
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2650
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_B.prj
Status: finished successfully.
Execution took 79.164377450943 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_F.prj
============================================================
Running simulation for load case: F with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
[mid=0] Solid density set from material: 2750
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2750
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2750
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_F.prj
Status: finished successfully.
Execution took 71.24448752403259 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
[mid=0] Solid density set from material: 2650
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2650
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2650
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_F.prj
Status: finished successfully.
Execution took 79.48885011672974 s
mesh path: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/mesh_GreatCell_fullFracture
[DEBUG] Current working directory: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/ogs/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook
[DEBUG] output project file: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_C.prj
============================================================
Running simulation for load case: C with method: LIE, tension_cutoff: true
============================================================
------------------------------------------------------------
* Running the simulation for sample Gneiss:
------------------------------------------------------------
[mid=0] Solid density set from material: 2750
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2750
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2750
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_C.prj
Status: finished successfully.
Execution took 70.04024767875671 s
------------------------------------------------------------
* Running the simulation for sample Greywacke:
------------------------------------------------------------
[mid=0] Solid density set from material: 2650
[mid=0] Bulk properties set
[mid=1] Solid density set from material: 2650
[mid=1] Bulk properties set
[mid=2] Solid density set from rubber: 1500
[mid=2] Bulk properties set
[mid=3] Solid density set from material: 2650
[mid=3], fracture, Bulk skipped
Project file written to output.
Simulation: /var/lib/gitlab-runner/builds/t1_R7hwLX/1/ogs/build/release-all/Tests/Data/LIE/HydroMechanics/GreatCelljupyterNotebook/GreatCellHM/HM2a_LIE_C.prj
Status: finished successfully.
Execution took 77.98217844963074 s
Post-processing
plotter = Plotter(…
(click to toggle)
plotter = Plotter(
output_dir=out_dir,
save_extracted_data=True,
)
data_dir = Path("external_data")
external_data = Plotter.load_external_data(data_dir, benchmark_tag="HM2a")
plotter.plot_volumetric_strain_vs_angle(
vtu_files_dict_full,
model_type="HM2a",
ylim_range=[-7.5, 2.5],
layout="subplots",
external_data=external_data["strain"],
)

plotter.plot_field_variables(vtu_files_dict_full)Load: A, File 0, Benchmark: HM2a, Material: Gneiss

Load: A, File 1, Benchmark: HM2a, Material: Greywacke

Load: E, File 0, Benchmark: HM2a, Material: Gneiss

Load: E, File 1, Benchmark: HM2a, Material: Greywacke

Load: B, File 0, Benchmark: HM2a, Material: Gneiss

Load: B, File 1, Benchmark: HM2a, Material: Greywacke

Load: F, File 0, Benchmark: HM2a, Material: Gneiss

Load: F, File 1, Benchmark: HM2a, Material: Greywacke

Load: C, File 0, Benchmark: HM2a, Material: Gneiss

Load: C, File 1, Benchmark: HM2a, Material: Greywacke

plotter.material_names = ["Gneiss", "Greywacke"]…
(click to toggle)
plotter.material_names = ["Gneiss", "Greywacke"]
plotter.vtu_file_names = {"LIE": vtu_files_dict_full}
plotter.plot_avg_width_vs_stress(
benchmark_tag="HM2a",
metric="width",
methods_to_include=["LIE"],
pee_load_values=PEE_load_values,
external_data=external_data["average"],
)
plotter.plot_avg_width_vs_stress(
benchmark_tag="HM2a",
metric="permeability",
methods_to_include=["LIE"],
pee_load_values=PEE_load_values,
external_data=external_data["average"],
)


array([ 1000000. , 2318019.48466054, 5500000. ,
8681980.51533946, 10000000. ])
lie_profiles = {}…
(click to toggle)
lie_profiles = {}
for load_case, file_list in plotter.vtu_file_names["LIE"].items():
lie_profiles[load_case] = plotter.extract_lie_aperture_from_list(
file_list,
plotter.material_names,
)
plotter.plot_fracture_aperture_profiles(
widthProfile=lie_profiles,
benchmark_tag="HM2a",
downsample=1,
# ylim=(0, 1e-5),
method_label="LIE",
external_data=external_data["widthProfile"],
)





pairs_to_check = {…
(click to toggle)
pairs_to_check = {
"HM1_HM_A_Greywacke": "HM1",
"HM2a_LIE_A_Greywacke": "HM2a",
"HM2b_LIE_A_Greywacke": "HM2b",
}
for case, label in pairs_to_check.items():
print(f"\n===== {label} case =====")
new_result = np.load(Path(out_dir, f"extracted_{case}_volStrain.npz"))
expected_result = np.load(Path("expected", f"extracted_{case}_volStrain.npz"))
eps_v_new = new_result["eps_v"]
eps_v_expected = expected_result["eps_v"]
phi_new = new_result["phi"]
phi_expected = expected_result["phi"]
np.testing.assert_allclose(eps_v_new, eps_v_expected, atol=7e-4)
np.testing.assert_allclose(phi_new, phi_expected, atol=1e-6)
print(f"\n{label} case passed.")===== HM1 case =====
HM1 case passed.
===== HM2a case =====
HM2a case passed.
===== HM2b case =====
HM2b case passed.
Reference
-
McDermott, C.I., Fraser-Harris, A., Sauter, M., Couples, G.D., Edlmann, K., Kolditz, O., Lightbody, A., Somerville, J. and Wang, W., 2018. New experimental equipment recreating geo-reservoir conditions in large, fractured, porous samples to investigate coupled thermal, hydraulic and polyaxial stress processes. Scientific reports, 8(1), p.14549.
-
Mollaali, M., Kolditz, O., Hu, M., Park, C.H., Park, J.W., McDermott, C.I., Chittenden, N., Bond, A., Yoon, J.S., Zhou, J. and Pan, P.Z., Liu H., Hou W., Lei H., Zhang L., Nagel T., Barsch M., Wang W., Nguyen S., Kwon S. and Yoshioka K., 2023. Comparative verification of hydro-mechanical fracture behavior: Task G of international research project DECOVALEX–2023. International Journal of Rock Mechanics and Mining Sciences, 170, p.105530.
-
Mollaali, M., Wang, W., You, T., Nagel, T., Fraser-Harris, A., McDermott, C., and Kolditz, O., 2025. Numerical benchmarking of GREAT cell experiments: Poly-axial stress effects on fluid flow in fractured rock using smeared and discrete methods.
This article was written by Mostafa Mollaali, Wenqing Wang. If you are missing something or you find an error please let us know.
Generated with Hugo 0.147.9
in CI job 635984
|
Last revision: July 16, 2025